Main Page: Difference between revisions
No edit summary |
No edit summary |
||
| Line 15: | Line 15: | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0002.png|120px]] | [[Image:LabelledCropped GPT Snapdragon 2010-000570-0002.png|120px]] | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0007.png|120px]] | [[Image:LabelledCropped GPT Snapdragon 2010-000570-0007.png|120px]] | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0003 double.png| | [[Image:LabelledCropped GPT Snapdragon 2010-000570-0003 double.png|100px]] | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0002 triple.png|120px]] | [[Image:LabelledCropped GPT Snapdragon 2010-000570-0002 triple.png|120px]] | ||
|} | |} | ||
| Line 21: | Line 21: | ||
The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 Kennaway et al 2011]). The GPT-framework was used to capture an understanding of (to model) the growing leaf (Kuchen et al 2012) and Snapdragon flower [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000537 Green et al 2011]. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000538 Cui et al 2010.]<br><br> | The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos ([http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 Kennaway et al 2011]). The GPT-framework was used to capture an understanding of (to model) the growing leaf (Kuchen et al 2012) and Snapdragon flower [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000537 Green et al 2011]. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000538 Cui et al 2010.]<br><br> | ||
[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:DarkGreen;">More details on growth </span>]]<br><br> | |||
==<span style="color:DarkGreen;">Viewing three dimensional images== | |||
{| border="0" width=100% style="background-color:#000000;" | {| border="0" width=100% style="background-color:#000000;" | ||
|- | |- | ||
|align="center"| | |align="center"| | ||
[[Image:Cs0prxz0.png|32x32px]] | [[Image:Cs0prxz0.png|32x32px]] | ||
[[Image:Leaf_trichomes.png|50px]] | |||
[[Image:Leaf_trichomes.png| | [[Image:Cs0prxz0.png|50px]] | ||
[[Image:GL2_GUS.png|50px]] | |||
[[Image:Leaf5.png|50px]] | |||
[[Image:OleosinSeed.png|50px]] | |||
[[Image:Cs0prxz0.png| | [[Image:OPT_Leaf_copy.png|50px]] | ||
[[Image:GL2_GUS.png| | [[Image:Seedling_copy.png|50px]] | ||
[[Image:Snapdragon_Peloric_mutant.png|50px]] | |||
[[Image:Leaf5.png| | [[Image:Tissue.png|50px]] | ||
[[Image:Z9r3j2yx.png|50px]] | |||
[[Image:OleosinSeed.png| | [[Image:1896_wh_txr_light.png|50px]] | ||
[[Image:OPT_Leaf_copy.png| | [[Image:Ara_flower.png|50px]] | ||
[[Image:Seedling_copy.png| | [[Image:Arableaf_ath8_OPT.png|50px]] | ||
[[Image:Snapdragon_Peloric_mutant.png| | |||
[[Image:Tissue.png| | |||
[[Image:Z9r3j2yx.png| | |||
[[Image:1896_wh_txr_light.png| | |||
[[Image:Ara_flower.png| | |||
[[Image:Arableaf_ath8_OPT.png| | |||
|} | |} | ||
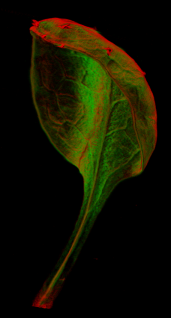
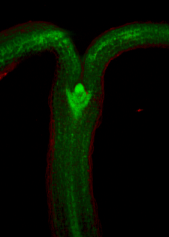
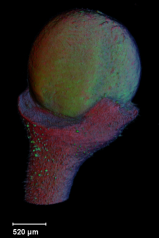
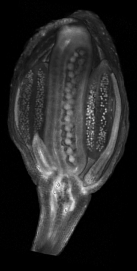
A collection of images of plants, plant organs and cells.<br><br> | |||
VolViewer uses [http://www.opengl.org/ OpenGL] and [http://qt.nokia.com/products/ Qt] to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. It is open-source and is also compatible with the Open Microscopy Environment ([http://openmicroscopy.org/site OME]).<br><br> | |||
[[Software#Viewing and measuring volume images: VolViewer|<span style="color:DarkGreen;">More details on viewing three dimensional images</span>]]<br><br> | |||
Revision as of 10:00, 21 May 2012
Bangham Lab - Home
Current activity: a collaboration with the CoenLab with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.
Computational biology toolboxes
Growing complex biological shapes from patterns of gene expression
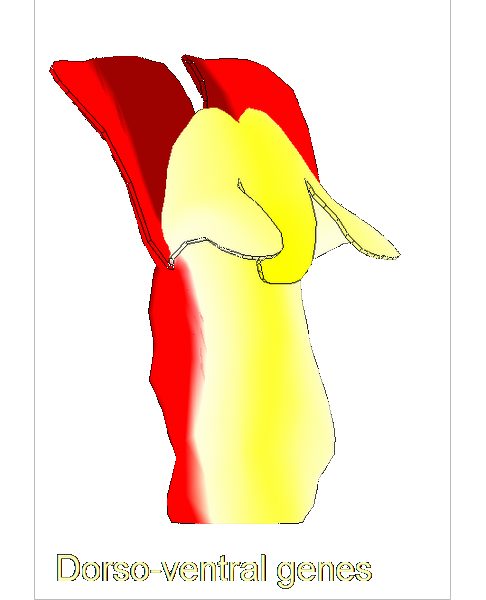
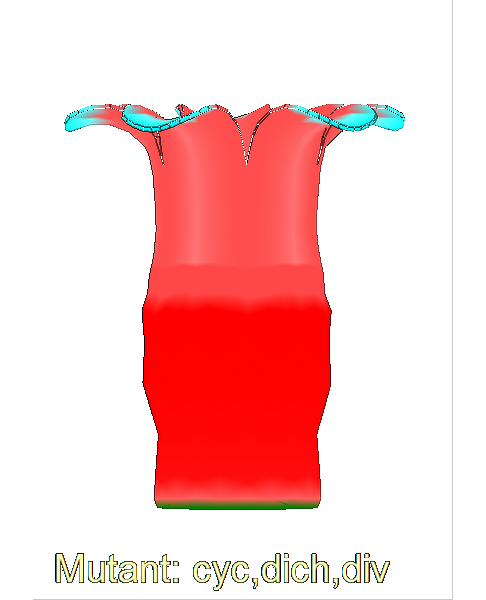
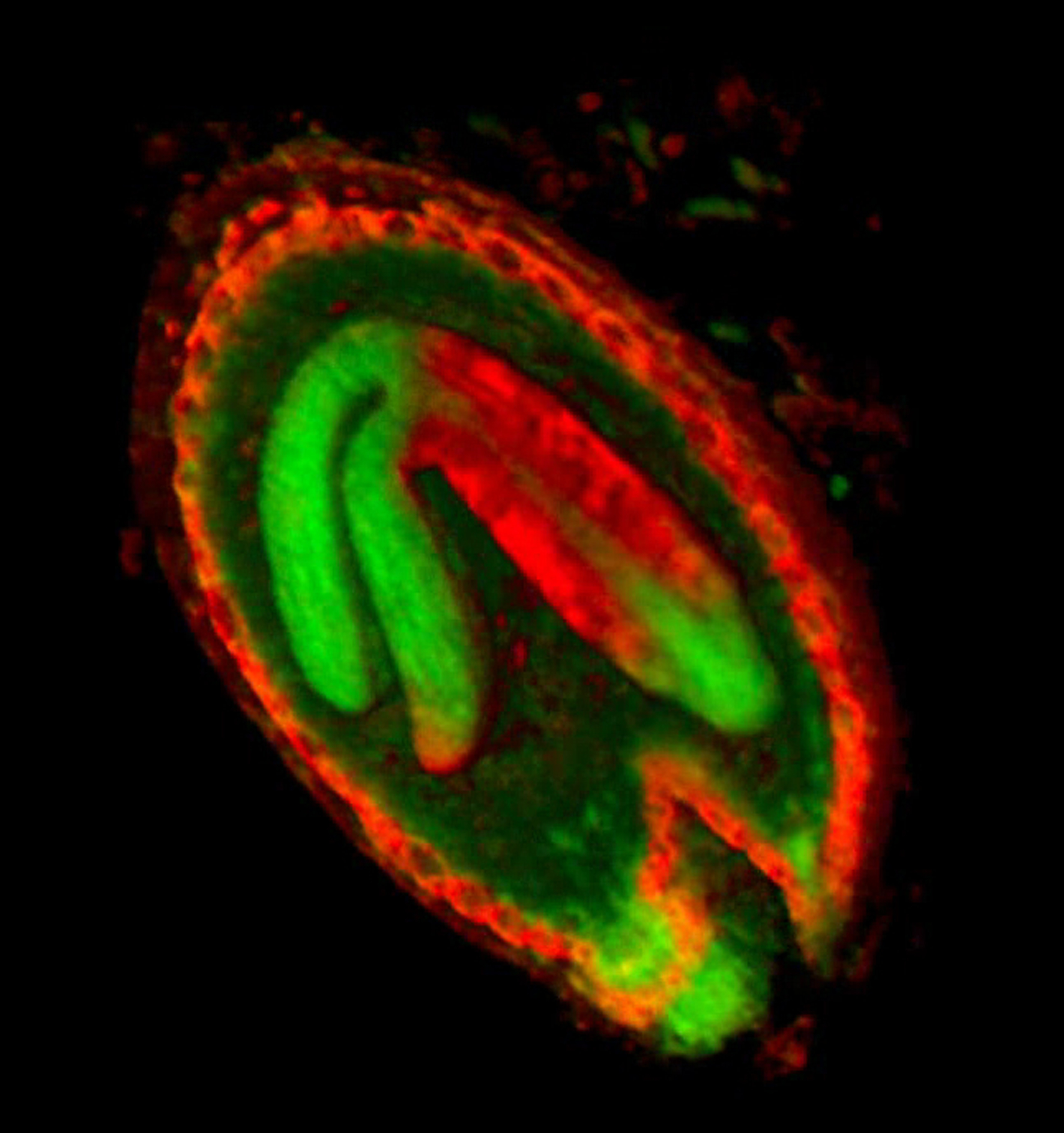
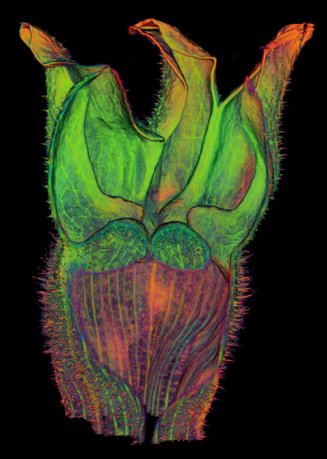
The growth of a complex snapdragon flower shape. Key to the model, is an hypothesis on how organisers control the axes along which growth occurs. The organisers are shown in cyan and green. On the right are the shapes of two symmetrical mutants computed from the same model (hypotheses).
The Growing Polarised Tissue Framework for understanding and modelling the relationship between gene activity and the growth of shapes such leaves, flowers and animal embryos (Kennaway et al 2011). The GPT-framework was used to capture an understanding of (to model) the growing leaf (Kuchen et al 2012) and Snapdragon flower Green et al 2011. The Snapdragon model was validated by comparing the results with other mutant and transgenic flowers Cui et al 2010.
Viewing three dimensional images
A collection of images of plants, plant organs and cells.
VolViewer uses OpenGL and Qt to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. It is open-source and is also compatible with the Open Microscopy Environment (OME).
More details on viewing three dimensional images
AAMToolbox
| <imgicon>AAMToolbox_logo.jpg|120px|AAMToolbox</imgicon> | For analysing populations of shapes and colours within the shapes using principal component analysis. What? How? Where? (PC, Mac, Linux, uses Matlab |
The AAMToolbox enables the user analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading, we have used it extensively for analysing the shapes of leaves and petals. The analysis can be applied to art, for example, finding systematic differences between portraits by, for example, Rembrandt and Modigliani. |