Research: Difference between revisions
JeromeAvondo (talk | contribs) No edit summary |
JeromeAvondo (talk | contribs) No edit summary |
||
| Line 19: | Line 19: | ||
='''Overview of Research Programme'''= | ='''Overview of Research Programme'''= | ||
Our research goal is to understand the algorithms underpinning the evolution and growth of plants. | |||
=='''Visualization, Quantification and Data Management of Microscopy Images'''== | =='''Visualization, Quantification and Data Management of Microscopy Images'''== | ||
Revision as of 11:37, 14 July 2010
|
Overview of Research Programme
Our research goal is to understand the algorithms underpinning the evolution and growth of plants.
Visualization, Quantification and Data Management of Microscopy Images

Jerome Avondo - email
The field of biological imaging has seen incredible advances due to new imaging techniques and molecular markers. These advances in data aquisition result in the creation of multidimensional images. How these images are to be visualized, quantified and data managed remains an open challenge.
To visualize and quantify multidimensional images I research the field of real time volume rendering and the use of the graphics processing unit (GPU). This allows the creation of rendering algorithms for the interactive visualization of microscopy images through space, spectral channels and time.
By using interactive visualization, it is also possible to create simple intuitive interfaces to equip biologists with tools to quantify their biological data. All results from my work is available to the community as a free open source application called VolViewer.
Due to the large amounts of data being produced by bio-image data acquisition systems, such as a confocal microscope, it is possible for a single lab to generate many terabytes of imaging data. To address the data management issues involved with such large data sets I am working in collaboration with the Open Microscopy Environment team lead by Jason Sweldow at the Univeristy of Dundee.
Together we are working on integrating the VolViewer application as an addon package to the Open Microscopy Environment platform. Work is also being conducted to offer a customized OMERO system as part of the John Innes Centre Bio-Imaging platform service. This work is being conducted to promote advanced visualisation, data management and image processing at the John Innes Centre. Research from this work can be found on the DMBI project wiki.
Modelling and analysis of growing plant tissues. Application to flower development
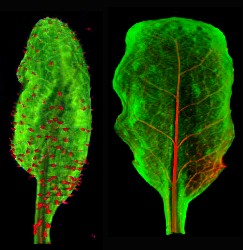
Pierre Barbier-De-Reuille - email
My main research project is about methods and tools for the modelling and the analysis of growing plant tissues.
For the modelling part, I am developing VVe, a modelling environment for the modelling of 2D structures in 3D space. VVe includes specialized tools for dealing with the development of plant tissues at the cellular scale, but can also be used to model tissue considered as a continuum. It is already distributed, as it is used by Sarah Robinson in our lab, in the lab of the professor Prusinkiewicz in Calgary and in the lab of the professor Kuhlemeier in Bern.
For the data analysis, I am developing digitizing and computation tools. I am in particular working on tracking cell growth and division, and extracting useful information from this (i.e. growth tensor, division rate, ...).
Currently, my developments aim at helping the understanding of the growth patterns in the mature petal of Arabidopsis.
Modelling microtubule dynamics
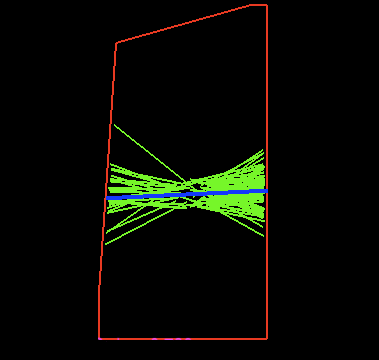
Scott Grandison - email
Microtubules are small, filamentous polymers that perform a variety of important developmental functions that have an impact, not only within the cell, but on a macroscopic scale. They help to provide structural integrity to the cell, by guiding the mechanisms which lay down cellulose microfibrils. If these microfibrils are laid down with a preferential direction then the cell can be encouraged to grow anisotropically which is important when forming complex shapes during development. Another important role for microtubules is their role in determining where exactly a cell will divide during mitosis.
I'm interesting in studying these functions by making accurate mathematical and computer simulations of microtubule dynamics. I work closely with Dr. Jordi Chan and Dr. Paul Southam who are interesting in the experimental and image processing part of the project.
Modelling Growth at the Tissue Scale
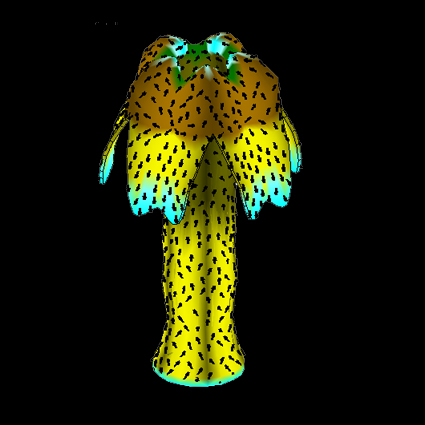
Richard Kennaway - email
I am developing finite element methods for modelling the growth and development of curved two-dimensional tissues such as leaves and petals. The interactive software tool I am developing, called GFtbox, is available for download.
Image Processing for Biological Development
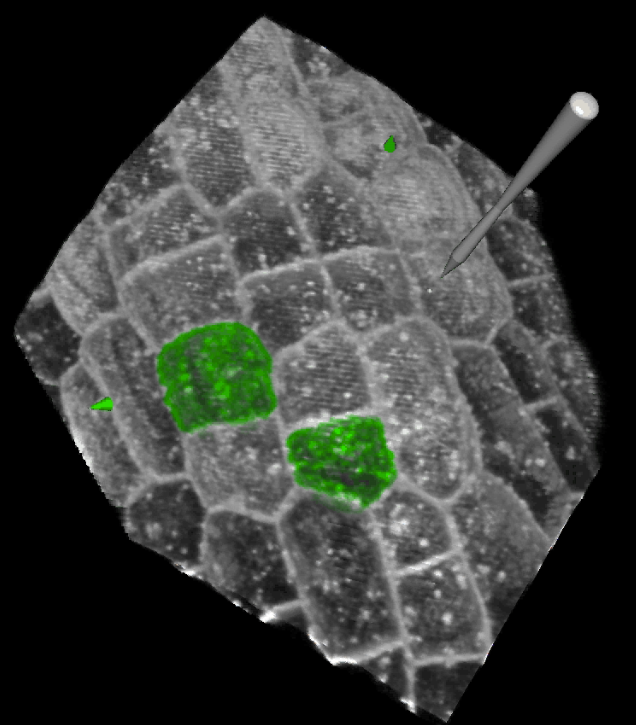
Paul Southam - email
Extracting quantitative measurements from image data is a fundamental step towards understanding how biological forms develop and evolve. I work on a number of projects including:
Modelling microtubule dynamics during growth and division - Jordi Chan & Scott Grandison.
Modelling Dynamic Growth Maps of Leaf Development - Samantha Fox.
Capturing plant development in 3D with Optical Projection Tomography - Karen Lee.
I am also interested in cell segmentation, cell registration, leaf classification and texture analysis.