VolViewer: Difference between revisions
JeromeAvondo (talk | contribs) No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Software| All open release software from the BanghamLab]]<br> | [[Software| All open release software from the BanghamLab]]<br> | ||
= | =What? How? Where?= | ||
[[Image:VolViewer.png|256px|thumb|The VolViewer main application window.]] | [[Image:VolViewer.png|256px|thumb|The VolViewer main application window.]] | ||
Revision as of 11:03, 9 February 2012
All open release software from the BanghamLab
What? How? Where?
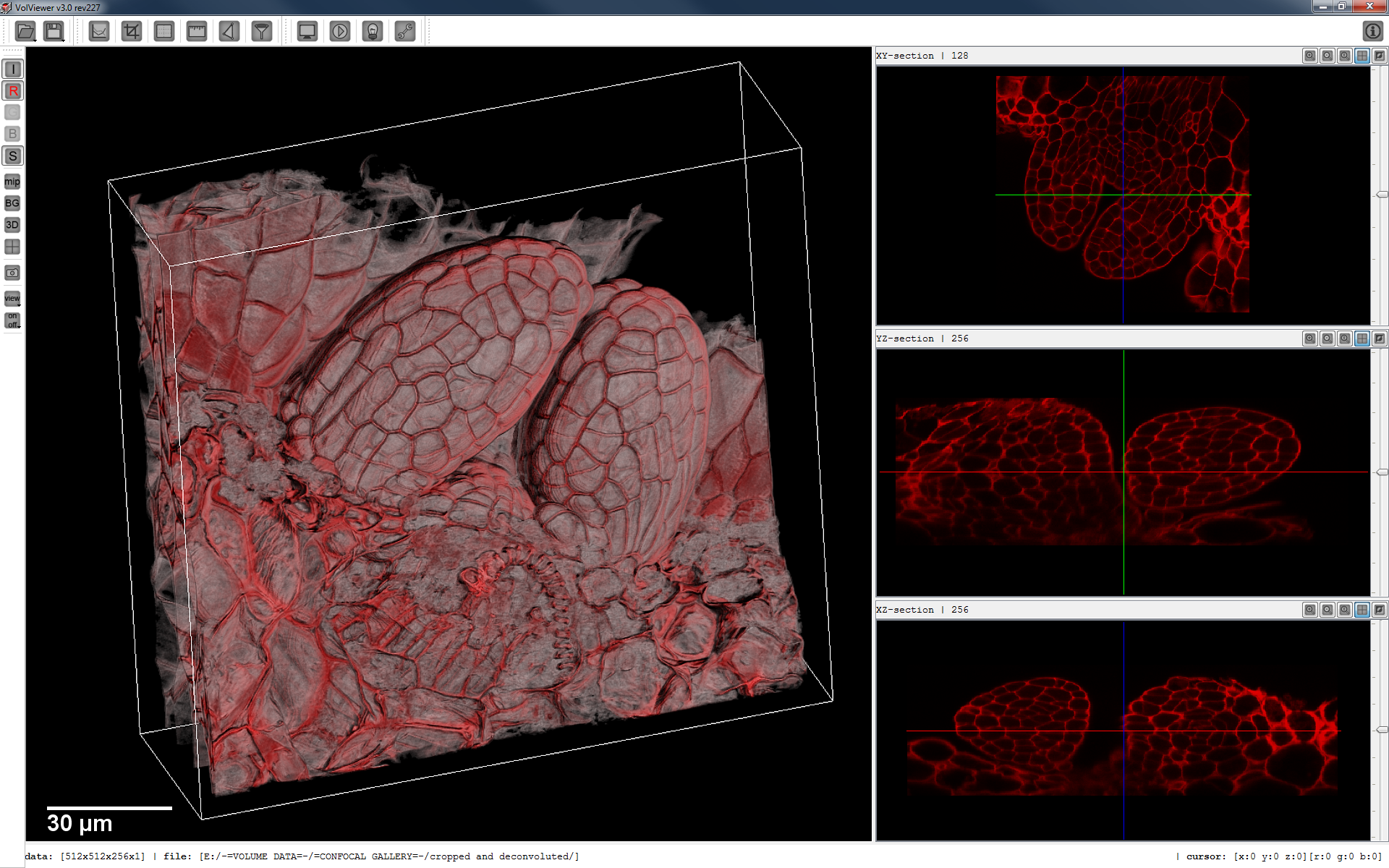
VolViewer is an open source application for the interactive visualisation and quantification of biological data. The application is written in C++ using OpenGL and Qt.
Features:
- Real-time volume rendering using an optimized 3D texture slicing algorithm.
- Interactive transfer functions to independently adjust opacity and intensity for up to three data channels.
- Real-time per channel thresholding, brightness and contrast operators.
- On-the-fly gradient computation for local illumination.
- Iso-surface computation with surface smoothing.
- Section viewing in any orientation / position.
- Real-time volume clipping.
- 3D measurements, filters & segmentation.
- Key frame interpolation for easy movie creation.
- Stereo rendering using either quad buffer or anaglyph mode.
Requirements: An OpenGL 2.1 / GLSL 1.20 compatible GPU with a recomended 512MB of memory.
User Documentation
- Tutorial on dealing with proprietory file formats (BioformatsConverter)
- Tutorial on loading and saving data
- Tutorial on user interface
- Tutorial on exploring data
- Tutorial on high quality projections and movies
- Tutorial on filters, quantification, segmentation and scripting
- Scripting in more detail - e.g. Matlab functions
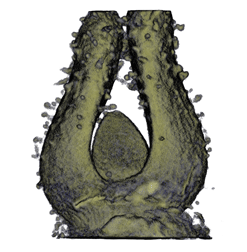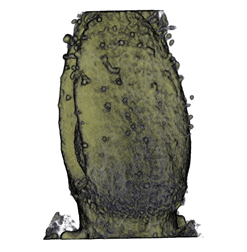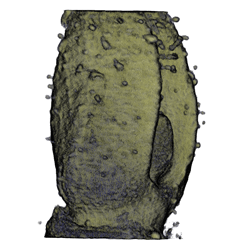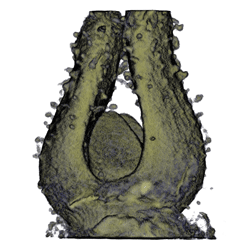
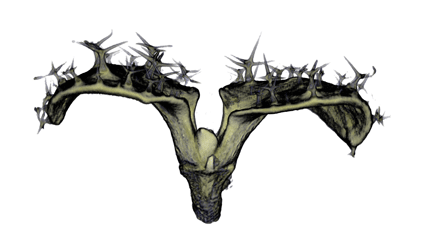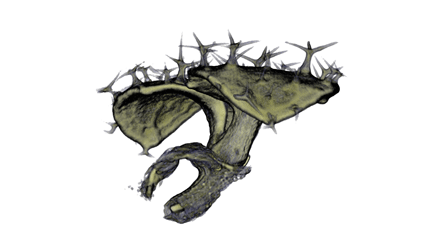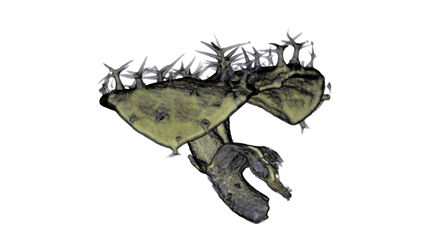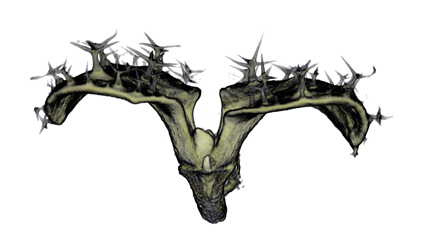
Sample Data
 |
 |
 |

|
| Antirinhium Meristem | Arabidopsis Seedling | Arabidopsis Leaf (GL2:GUS expression in red) | Arabidopsis Leaf (Ath8:::GUS expression in red) |
| Download | Download | Download | Download |
* all data courtesy of Karen Lee [1]
Download
Although we try to keep up to date builds these sometimes lag behind the SVN trunk. So if you want the latest version / features, it is best to build the application from the trunk of the SVN. The build system is based on qmake for easy cross platform compilation.
| Windows (32bit) | Windows (64bit) | Linux | MacOS X (i386/x86_64/10.5+) |
Note that for the windows versions you will need to install the corresponding Microsoft Visual C++ 2010 SP1 Redistributable Package which can be found here: 32bit and 64bit.
Source Code
Public SVN: https://cmpdartsvr1.cmp.uea.ac.uk/banghamlabSVN/VolViewer/
Building from source
Media/Press
VolViewer has appeared in the following:
Front cover: Handbook of Plant Science | Front cover: The Plant Cell | Royal Microscopical Society: Infocus Magazine | Bundled with the Bioptonic 3001 scanner: Bioptonics Viewer | The Guardian newspaper: 3D Fruit fly | Qt Ambassador program | Triffid Nurseries website