Details on hydrophobicity plots: Difference between revisions
No edit summary |
|||
| Line 12: | Line 12: | ||
Actually, for the data-sieve I did use a cascade of medians which was a very inefficient algorithm. Subsequently, I switched to a cascade of 1D recursive medians which also preserved scale space but was hugely quicker and is idempotent. | Actually, for the data-sieve I did use a cascade of medians which was a very inefficient algorithm. Subsequently, I switched to a cascade of 1D recursive medians which also preserved scale space but was hugely quicker and is idempotent. | ||
[[Image:hydrophobicity_plots_Fasman.jpg|700px]] | [[Image:hydrophobicity_plots_Fasman.jpg|700px]]<br> | ||
From Fasman and Gilbert "The prediction of transmembrane protein sequences and their conformation: an evaluation" in Trends in Biochemistry 15 pp 89:91 | From Fasman and Gilbert "The prediction of transmembrane protein sequences and their conformation: an evaluation" in Trends in Biochemistry 15 pp 89:91 | ||
Left: Running mean (c.f Gaussian): no good, Right: Median based Data-sieve: much better<br><br> | Left: Running mean (c.f Gaussian): no good, Right: Median based Data-sieve: much better<br><br> | ||
These authors compared many different methods and concluded that the data-sieve was best. However, they found a reason not to recommend it: that they didn't understand how it worked.<br><br> | These authors compared many different methods and concluded that the data-sieve was best. However, they found a reason not to recommend it: that they didn't understand how it worked.<br><br> | ||
Well there you are then. Means verses medians.<br><br> | Well there you are then. Means verses medians.<br><br> | ||
The good thing was that I was motivated to prove exactly how it worked and why it was so good - I moved to Computing Sciences and published everything in computing journals. I was also so excited about the discovery of a new set of transforms that could underpin brain vision that I attended conferences on vision - Horace Barlows retirement bash, etc. All I got from, for example, distinguished scientists like Colin Blakemore was: 'But mathematicians have shown that Gaussian Filter banks are unique ...'<br><br. | The good thing was that I was motivated to prove exactly how it worked and why it was so good - I moved to Computing Sciences and published everything in computing journals. I was also so excited about the discovery of a new set of transforms that could underpin brain vision that I attended conferences on vision - Horace Barlows retirement bash, etc. All I got from, for example, distinguished scientists like Colin Blakemore was: 'But mathematicians have shown that Gaussian Filter banks are unique ...'<br><br>. | ||
I was proving that they were not unique. They are unique among linear filters but not filters in general. | I was proving that they were not unique. They are unique among linear filters but not filters in general. | ||
Revision as of 19:03, 15 June 2014
MSER_and_Sieve_Details#ApplicationsBack to MSERs etc
Trying to spot regions (lengths) that are up (hydrophobic) and down (hydrophilic)
Left: Running mean (c.f Gaussian): no good, Right: Median based Data-sieve: much better
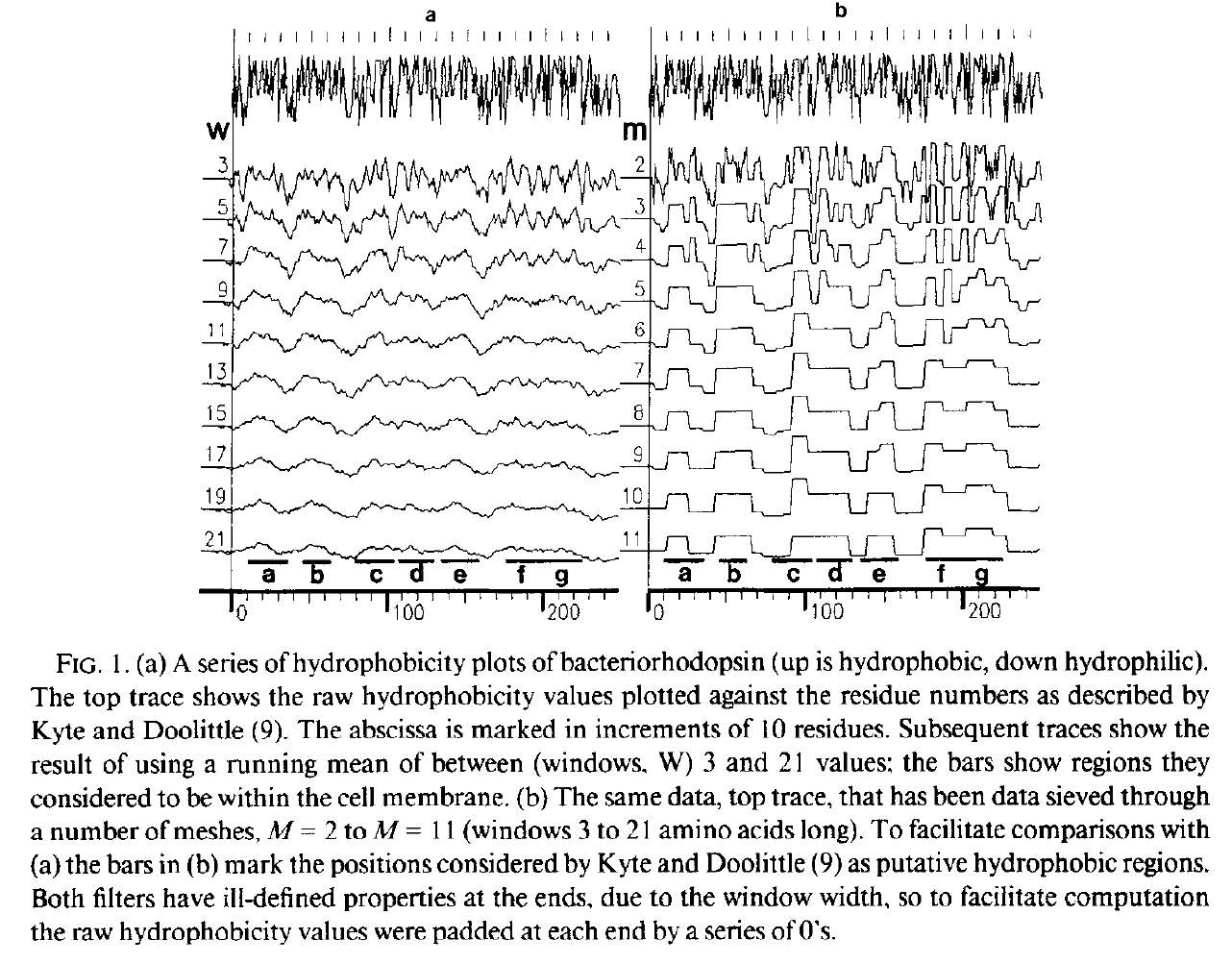 Bangham, J.A. (1988) Data-sieving hydrophobicity plots. Anal. Biochem. 174, 142–145
Bangham, J.A. (1988) Data-sieving hydrophobicity plots. Anal. Biochem. 174, 142–145
Abstract from ...
Hydrophobicity plots provide clues to the tertiary structure of proteins (J. Kyte and R. F. Doolittle, 1982, J. Mol. Biol.157, 105; C. Chothia, 1984, Annu. Rev. Biochem.53, 537; T. P. Hopp and K. R. Woods, 1982, Proc. Natl. Acad. Sci. USA78, 3824). To render domains more visible, the raw data are usually smoothed using a running mean of between 5 and 19 amino acids. This type of smoothing still incorporates two disadvantages. First, peculiar residues that do not share the properties of most of the amino acids in the domain may prevent its identification. Second, as a low-pass frequency filter the running mean smoothes sudden transitions from one domain, or phase, to another. Data-sieving is described here as an alternative method for identifying domains within amino acid sequences. The data-sieve is based on a running median and is characterized by a single parameter, the mesh size, which controls its resolution. It is a technique that could be applied to other series data and, in multidimensions, to images in the same way as a median filter.
Actually, for the data-sieve I did use a cascade of medians which was a very inefficient algorithm. Subsequently, I switched to a cascade of 1D recursive medians which also preserved scale space but was hugely quicker and is idempotent.
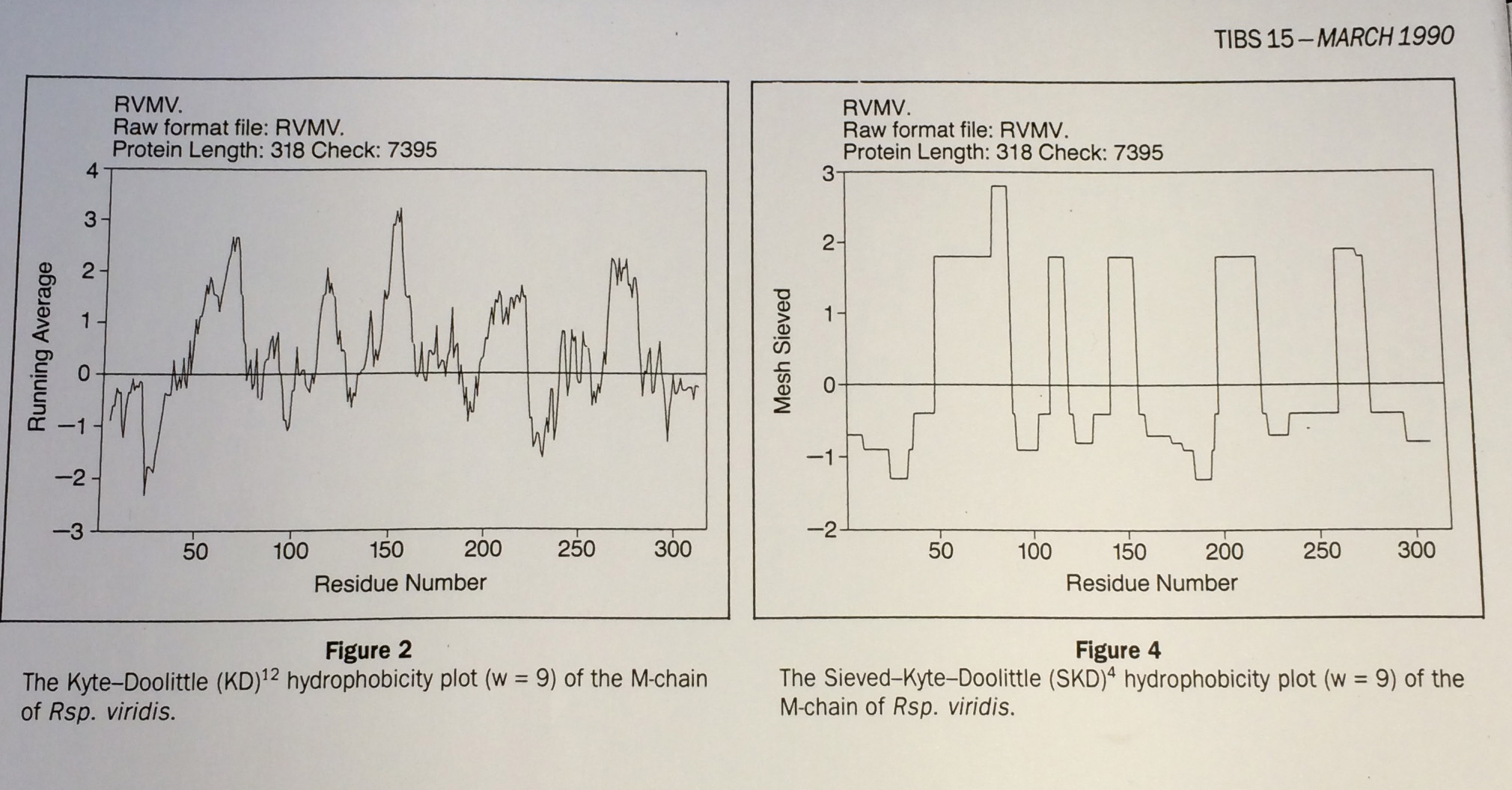
From Fasman and Gilbert "The prediction of transmembrane protein sequences and their conformation: an evaluation" in Trends in Biochemistry 15 pp 89:91
Left: Running mean (c.f Gaussian): no good, Right: Median based Data-sieve: much better
These authors compared many different methods and concluded that the data-sieve was best. However, they found a reason not to recommend it: that they didn't understand how it worked.
Well there you are then. Means verses medians.
The good thing was that I was motivated to prove exactly how it worked and why it was so good - I moved to Computing Sciences and published everything in computing journals. I was also so excited about the discovery of a new set of transforms that could underpin brain vision that I attended conferences on vision - Horace Barlows retirement bash, etc. All I got from, for example, distinguished scientists like Colin Blakemore was: 'But mathematicians have shown that Gaussian Filter banks are unique ...'
.
I was proving that they were not unique. They are unique among linear filters but not filters in general.