MSER's and Connected sets: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 1: | Line 1: | ||
[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#MSERs.2C_extrema.2C_connected-set_filters_and_sieves Return to connected-sets]<br><br> | [http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#MSERs.2C_extrema.2C_connected-set_filters_and_sieves Return to connected-sets]<br><br> | ||
{| border="0" width=100% style="background-color:#ffffff;" | |||
|- | |||
|align="left"| | |||
[[Details_on_hydrophobicity_plots | The first published application of the data-sieve as reviewed by Fasman, click here ...</span>]] | |||
|[[Image:hydrophobicity_plots_Fasman.jpg|right|300px]] | |||
|From Fasman and Gilbert "The prediction of transmembrane protein sequences and their conformation: an evaluation" in Trends in Biochemistry 15 pp 89:91 | |||
Left: Running mean (c.f Gaussian): <span style="color:#C364C5;">no good</span>, Right: Median based Data-sieve: <span style="color:#C364C5;">much better</span> | |||
|} | |||
=<span style="color:Chocolate">'''''Extrema''''' in one dimensional signals<span style="color:Chocolate">= | =<span style="color:Chocolate">'''''Extrema''''' in one dimensional signals<span style="color:Chocolate">= | ||
====One dimensional A==== | ====One dimensional A==== | ||
Revision as of 17:51, 30 July 2014
|
The first published application of the data-sieve as reviewed by Fasman, click here ... |
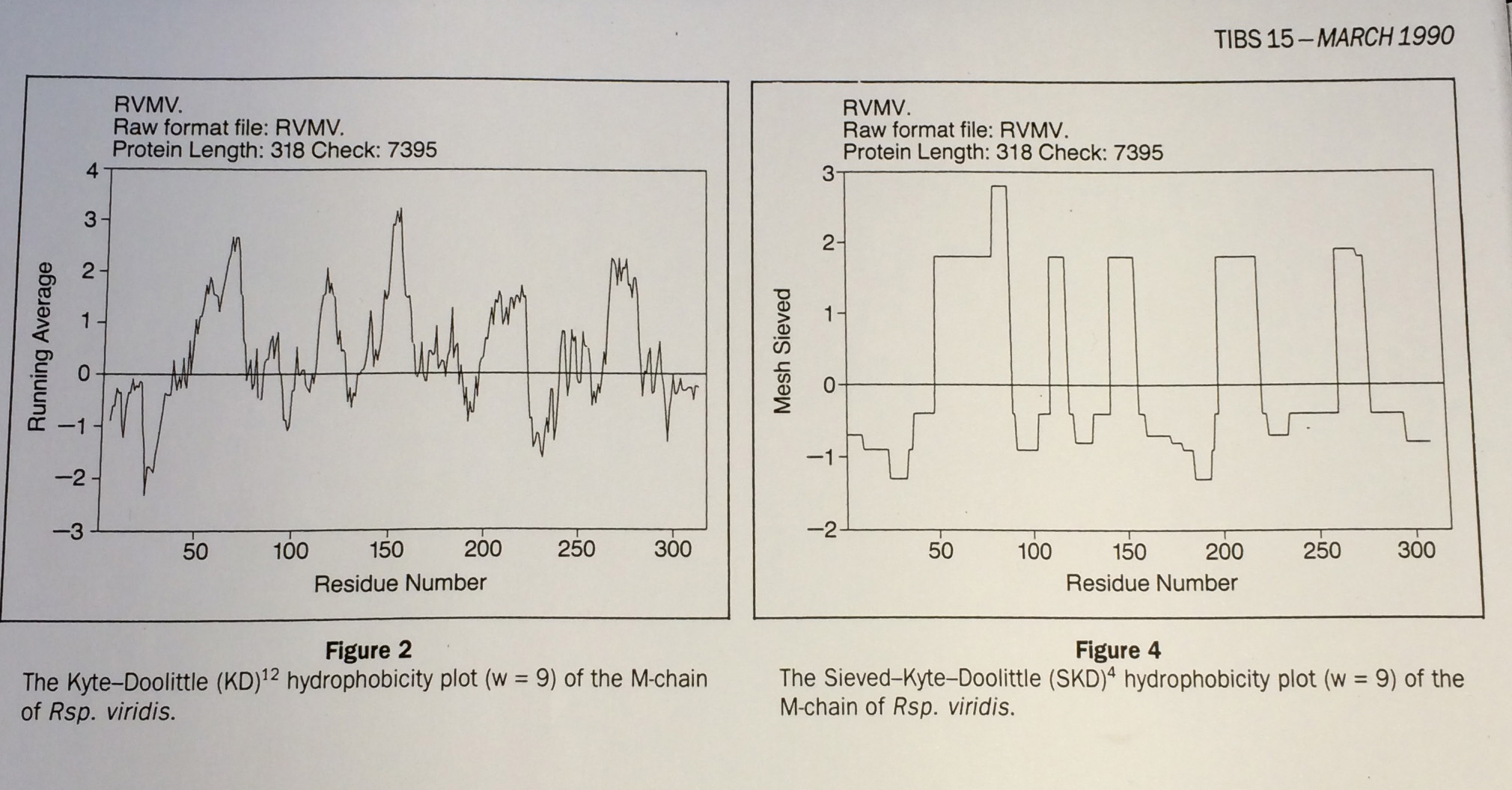 |
From Fasman and Gilbert "The prediction of transmembrane protein sequences and their conformation: an evaluation" in Trends in Biochemistry 15 pp 89:91
Left: Running mean (c.f Gaussian): no good, Right: Median based Data-sieve: much better |
Extrema in one dimensional signals
One dimensional A
| What are MSER's in one dimension? | 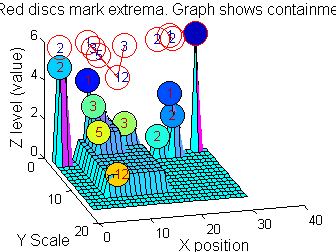
|
One dimensional B
| One dimensional extrema in images | 
|