GFtbox: Difference between revisions
No edit summary |
No edit summary |
||
| (50 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[ | [[Software#GFtbox|Back to BanghamLab software]]<br> | ||
==What is ''GFtbox''?== | [[GFtbox Tutorial pages|''Tutorials'']] and [https://sourceforge.net/p/gftbox/ ''Download'']<br> | ||
==<span style="color:Navy;">What is ''GFtbox'', how does it work and where is it?</span>== | |||
[[Image:GPT_thumbnail2.png|thumb|left|100px]] | [[Image:GPT_thumbnail2.png|thumb|left|100px]] | ||
'''We wish to understand''' how patterns of gene activity in biological organs influence the developing shape. Our suggestion is that genes may regulate growth direction independently of growth rate. We formalised our ideas in the Growing Polarised Tissue Framework ( | '''We wish to understand''' how patterns of gene activity in biological organs influence the developing shape. Our suggestion is that genes may regulate growth direction independently of growth rate. We formalised our ideas in the Growing Polarised Tissue Framework (reference 1, below). The icon is based on Fig. 4 of reference 2, and shows an asymmetrical spur that has grown under the influence of a polariser gradient, indicated by the arrows. | ||
'''Intuition is not''' a good guide to the behaviour of tissue growing under the influence of patterns of growth. Patterns interact with each other and the continuous sheet of tissue to produce complex shapes. It is therefore necessary to translate intuition into a mathematical model whose behaviour can be calculated. ''GFtbox'' is a package for creating and simulating such models. | |||
''' | '''A ''GFtbox'' project comprises two parts''', the Mesh and the Interaction function. The Mesh is a finite element model of the tissue, together with patterns of gene activity, tissue identity, and biochemical properties. The Interaction function specifies the local interactions of these properties. | ||
''' | '''Does ''GFtbox'' work correctly?''' Validation is an important question to be asked of all modelling software. There are various simple models for which analytical solutions can be obtained and compared with the computations performed by GFtbox. An extensive range of such validation tests can be found in Supplemental Text 1 of reference 2. | ||
We have used the '''GPT-framework''' to understand the development of the Snapdragon flower (reference 3 below). '''Using ''GFtbox''''' we started with a crown shaped sheet of tissue (the canvas), laid out observed or hypothesised patterns of regulatory activity and then grew the canvas in 3D. Patterning can continue during growth and the final shape was compared quantitatively with its biological counterpart -- so '''testing the hypotheses''' made in the model (reference 4 below). | |||
'''To work with ''GFtbox''''' is to practice thinking quantitatively about the relationship between genes, growth and form. | '''To work with ''GFtbox''''' is to practice thinking quantitatively about the relationship between genes, growth and form. | ||
==What does ''GFtbox'' require?== | |||
'''''GFtbox'' is written in [http://en.wikipedia.org/wiki/MATLAB Matlab].''' | <big>References</big><br> | ||
(* members of the biological computing BanghamLab) | |||
<ol> | |||
<li><small> Susanna Sauret-Güeto, Katharina Schiessl, Andrew Bangham*, Robert Sablowski, Enrico Coen (2013). JAGGED Controls Arabidopsis Petal Growth and Shape by Interacting with a Divergent Polarity Field PLOS Biology, 11 (4) : 10.1371/journal.pbio.1001550, [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1001550 Sauret-Güeto et al 2013]</small> | |||
<li><small> Erika E. Kuchen, Samantha Fox, Pierre Barbier de Reuille*, Richard Kennaway*, Sandra Bensmihen, Jerome Avondo*, Grant M. Calder, Paul Southam*, Sarah Robinson, Andrew Bangham*, Enrico Coen, 2012 "Generation of Leaf Shape through Early Patterns of Growth and Tissue Polarity",[http://www.sciencemag.org/content/335/6072/1092.abstract ''' Science''': 2 March 2012: 1092-1096.]</small> | |||
<li><small> Kennaway* R, Coen E, Green A, Bangham* A, 2011 Generation of Diverse Biological Forms through Combinatorial Interactions between Tissue Polarity and Growth. [http://www.ploscompbiol.org/article/info:doi/10.1371/journal.pcbi.1002071 PLoS Comput Biol 7(6): e1002071.]</small> | |||
<li><small> Amelia Green, Richard Kennaway*, Andrew Hanna, Andrew Bangham*, Enrico Coen, "Genetic Control of Organ Shape and Tissue Polarity", [http://dx.doi.org/10.1371/journal.pbio.1000537 PLoS Biol, 2010, vol.8, no.11.]</small> | |||
<li><small> Min-Long Cui, Lucy Copsey, Amelia Green, Andrew Bangham*, Enrico Coen, "Quantitative Control of Organ Shape by Combinatorial Gene Activity", [http://dx.doi.org/10.1371/journal.pbio.1000538 PLoS Biol, 2010, vol.8, no.11.]</small> | |||
</ol> | |||
==<span style="color:Navy;">What does ''GFtbox'' require?</span>== | |||
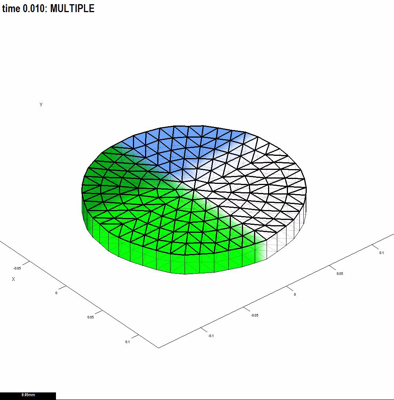
'''''GFtbox'' is written in [http://en.wikipedia.org/wiki/MATLAB Matlab].''' It does not require any extra Mathworks toolboxes, nor any separately compiled modules. Matlab is available as a [http://www.mathworks.com/products/matlab/tryit.html 30 day free trial] and as a [http://www.mathworks.com/academia/student_version/?s_cid=global_nav student edition]. ''GFtbox'' comprises around 1000 subroutines and 90,000 lines of code. It allows individual models, the part that a user writes, to be very compact -- 10 to 100 lines of Matlab code. For example, the model shown in Fig. 1 has a mesh that was created using the graphical user interface, and this code in its interaction function: | |||
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. | if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. | ||
id_a_p(m.nodes(:,1)<-0.03)=1; % | id_a_p(m.nodes(:,1) < -0.03) = 1; % set up region for A where identity factor A is represented by id_a_p | ||
id_b_p(m.nodes(:,2)<-0.01)=1; % | id_b_p(m.nodes(:,2) < -0.01) = 1; % set up region for B | ||
else | else | ||
% @@KRN Growth Regulatory Network | % @@KRN Growth Regulatory Network | ||
| Line 30: | Line 49: | ||
||<wikiflv width="352" height="288" logo="false" loop="true">GPT_why_matlab-2011-05-05-0005.flv|GPT_why_matlab-2011-05-05-0005.png</wikiflv> | ||<wikiflv width="352" height="288" logo="false" loop="true">GPT_why_matlab-2011-05-05-0005.flv|GPT_why_matlab-2011-05-05-0005.png</wikiflv> | ||
|-align="top" | |-align="top" | ||
|<center>Fig. 1. Mesh with two intersecting patterns of growth factors. Blue is factor A, Green factor B</center>||<center> | |<center>Fig. 1. Mesh with two intersecting patterns of growth factors. Blue is factor A, Green factor B</center>||<center>A movie of the mesh growing and subdividing. The intensity of red represents the growth rate</center> | ||
|} | |} | ||
'''Why Matlab?''' The language suits our problem - the | '''Why Matlab?''' The language suits our problem - the reasoning goes like this. Tissue is represented by a thin 3D mesh. Growth factors levels vary spatially forming patterns, e.g. Fig. 1. Here there are two, ''A'' and ''B''. We might make the hypothesis that the growth rate is specified by ''A'' but partially inhibited by ''B'' (inhibited by an amount ''K''). This is a simple idea that can be expressed in Matlab equally simply by writing ''Growth=A .* inh(K, B)''. This is because variables ''A'' and ''B'' can represent vectors - in this case a level for each node in the mesh. We also define a general inhibition function (''inh''). It means that it is straightforward to convert our thoughts on the biology into a programmatic description of a computational model. Moreover, the language is well documented with lots of convenient tools. In particular, Matlab has an extensive library of portable graphical user interface (GUI) functions - and this is convenient for producing tools to visualise the mesh and patterns of growth factors. | ||
'''The computational bottleneck''' in ''GFtbox'' is solving the finite element equations. We find that the Matlab [http://www.mathworks.com/help/techdoc/ref/cgs.html cgs] function is quicker and more accurate than alternatives from other sources (e.g. [http://software.intel.com/en-us/articles/intel-mkl/ lsqr], [http://www.culatools.com/forums/viewtopic.php?f=14&t=721 culaSgesvd]). The speed of ''GFtbox'' appears to be limited by memory-cpu bandwidth rather than cpu speed. It runs best interactively on a 64 bit machine with a large memory. | |||
Growing ''GFtbox'' models does not require the GUI. Everything done by the GUI can also be done at the Matlab command prompt or within a Matlab script file, Multiple models can be run as Matlab scripts on a computing cluster, and the results examined visually in the GUI. | |||
Matlab is sufficiently flexible that inexperienced programmers (and half our team are biologists, not programmers) can pick it up on-the-fly. It is an interpreted language, which makes it easy to step through code to follow what is happening ni case of difficulty. | |||
'''Matlab runs on''' Windows, Mac and Linux | '''Matlab runs on''' Windows, Mac and Linux. The key Matlab numerical algorithms are documented and generally visible and there is a large user-base providing lots of web based help. | ||
==How to start using ''GFtbox''== | ==<span style="color:Navy;">How to start using ''GFtbox''</span>== | ||
[http://sourceforge.net/p/gftbox/home/ '''''GFtbox'' can be freely downloaded''' from sourceforge]. After downloading, start Matlab, cd to the GFtbox directory, and give the command <code>GFtbox</code>. GFtbox will start and present you with its graphic interface. | |||
GFtbox automatically adds all of its subdirectories to the Matlab command path. If you give a <code>savepath</code> command at the Matlab console, then Matlab will remember where GFtbox is, and on subsequent runs of Matlab you will not have to cd to the GFtbox directory before running it. | GFtbox automatically adds all of its subdirectories to the Matlab command path. If you give a <code>savepath</code> command at the Matlab console, then Matlab will remember where GFtbox is, and on subsequent runs of Matlab you will not have to cd to the GFtbox directory before running it. | ||
'''In addition to the tutorials | [[GFtbox Tutorial pages|'''In addition to the tutorials given here''']], GFtbox comes with several example projects and the projects behind our published Figures. These projects appear on the Projects menu of the GFtbox window, in the Models submenu. There is a Help menu and when a new project is formed - the new associated interaction function is filled with comments that illustrate how the model can be written. In the GUI, mouseover text is provided for each of the GUI functions and the last item in every menu is a local help submenu. Documentation files can be found in the ''doc'' subdirectory of GFtbox. | ||
[[GFtbox Tutorial pages|'''Tutorial pages''']] | [[GFtbox Tutorial pages|'''Tutorial pages''']] | ||
Latest revision as of 16:30, 22 May 2013
Back to BanghamLab software
Tutorials and Download
What is GFtbox, how does it work and where is it?
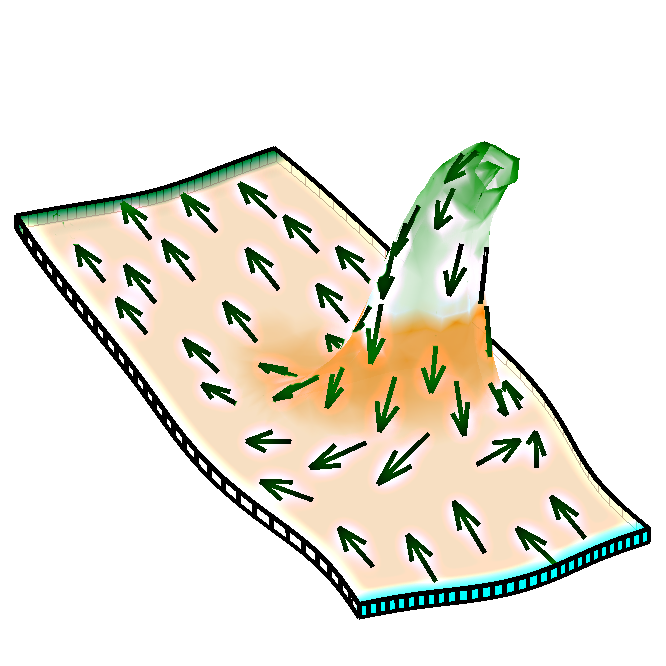
We wish to understand how patterns of gene activity in biological organs influence the developing shape. Our suggestion is that genes may regulate growth direction independently of growth rate. We formalised our ideas in the Growing Polarised Tissue Framework (reference 1, below). The icon is based on Fig. 4 of reference 2, and shows an asymmetrical spur that has grown under the influence of a polariser gradient, indicated by the arrows.
Intuition is not a good guide to the behaviour of tissue growing under the influence of patterns of growth. Patterns interact with each other and the continuous sheet of tissue to produce complex shapes. It is therefore necessary to translate intuition into a mathematical model whose behaviour can be calculated. GFtbox is a package for creating and simulating such models.
A GFtbox project comprises two parts, the Mesh and the Interaction function. The Mesh is a finite element model of the tissue, together with patterns of gene activity, tissue identity, and biochemical properties. The Interaction function specifies the local interactions of these properties.
Does GFtbox work correctly? Validation is an important question to be asked of all modelling software. There are various simple models for which analytical solutions can be obtained and compared with the computations performed by GFtbox. An extensive range of such validation tests can be found in Supplemental Text 1 of reference 2.
We have used the GPT-framework to understand the development of the Snapdragon flower (reference 3 below). Using GFtbox we started with a crown shaped sheet of tissue (the canvas), laid out observed or hypothesised patterns of regulatory activity and then grew the canvas in 3D. Patterning can continue during growth and the final shape was compared quantitatively with its biological counterpart -- so testing the hypotheses made in the model (reference 4 below).
To work with GFtbox is to practice thinking quantitatively about the relationship between genes, growth and form.
References
(* members of the biological computing BanghamLab)
- Susanna Sauret-Güeto, Katharina Schiessl, Andrew Bangham*, Robert Sablowski, Enrico Coen (2013). JAGGED Controls Arabidopsis Petal Growth and Shape by Interacting with a Divergent Polarity Field PLOS Biology, 11 (4) : 10.1371/journal.pbio.1001550, Sauret-Güeto et al 2013
- Erika E. Kuchen, Samantha Fox, Pierre Barbier de Reuille*, Richard Kennaway*, Sandra Bensmihen, Jerome Avondo*, Grant M. Calder, Paul Southam*, Sarah Robinson, Andrew Bangham*, Enrico Coen, 2012 "Generation of Leaf Shape through Early Patterns of Growth and Tissue Polarity", Science: 2 March 2012: 1092-1096.
- Kennaway* R, Coen E, Green A, Bangham* A, 2011 Generation of Diverse Biological Forms through Combinatorial Interactions between Tissue Polarity and Growth. PLoS Comput Biol 7(6): e1002071.
- Amelia Green, Richard Kennaway*, Andrew Hanna, Andrew Bangham*, Enrico Coen, "Genetic Control of Organ Shape and Tissue Polarity", PLoS Biol, 2010, vol.8, no.11.
- Min-Long Cui, Lucy Copsey, Amelia Green, Andrew Bangham*, Enrico Coen, "Quantitative Control of Organ Shape by Combinatorial Gene Activity", PLoS Biol, 2010, vol.8, no.11.
What does GFtbox require?
GFtbox is written in Matlab. It does not require any extra Mathworks toolboxes, nor any separately compiled modules. Matlab is available as a 30 day free trial and as a student edition. GFtbox comprises around 1000 subroutines and 90,000 lines of code. It allows individual models, the part that a user writes, to be very compact -- 10 to 100 lines of Matlab code. For example, the model shown in Fig. 1 has a mesh that was created using the graphical user interface, and this code in its interaction function:
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code.
id_a_p(m.nodes(:,1) < -0.03) = 1; % set up region for A where identity factor A is represented by id_a_p
id_b_p(m.nodes(:,2) < -0.01) = 1; % set up region for B
else
% @@KRN Growth Regulatory Network
kapar_p(:) = id_a_l .* inh(1,id_b_l); % growth rate
kaper_p(:) = kapar_p; % isotropic growth
kbpar_p(:) = kapar_p; % same on both sides of the sheet
kbper_p(:) = kapar_p; % same
knor_p(:) = 0; % thickness not growing
end
Why Matlab? The language suits our problem - the reasoning goes like this. Tissue is represented by a thin 3D mesh. Growth factors levels vary spatially forming patterns, e.g. Fig. 1. Here there are two, A and B. We might make the hypothesis that the growth rate is specified by A but partially inhibited by B (inhibited by an amount K). This is a simple idea that can be expressed in Matlab equally simply by writing Growth=A .* inh(K, B). This is because variables A and B can represent vectors - in this case a level for each node in the mesh. We also define a general inhibition function (inh). It means that it is straightforward to convert our thoughts on the biology into a programmatic description of a computational model. Moreover, the language is well documented with lots of convenient tools. In particular, Matlab has an extensive library of portable graphical user interface (GUI) functions - and this is convenient for producing tools to visualise the mesh and patterns of growth factors.
The computational bottleneck in GFtbox is solving the finite element equations. We find that the Matlab cgs function is quicker and more accurate than alternatives from other sources (e.g. lsqr, culaSgesvd). The speed of GFtbox appears to be limited by memory-cpu bandwidth rather than cpu speed. It runs best interactively on a 64 bit machine with a large memory.
Growing GFtbox models does not require the GUI. Everything done by the GUI can also be done at the Matlab command prompt or within a Matlab script file, Multiple models can be run as Matlab scripts on a computing cluster, and the results examined visually in the GUI.
Matlab is sufficiently flexible that inexperienced programmers (and half our team are biologists, not programmers) can pick it up on-the-fly. It is an interpreted language, which makes it easy to step through code to follow what is happening ni case of difficulty.
Matlab runs on Windows, Mac and Linux. The key Matlab numerical algorithms are documented and generally visible and there is a large user-base providing lots of web based help.
How to start using GFtbox
GFtbox can be freely downloaded from sourceforge. After downloading, start Matlab, cd to the GFtbox directory, and give the command GFtbox. GFtbox will start and present you with its graphic interface.
GFtbox automatically adds all of its subdirectories to the Matlab command path. If you give a savepath command at the Matlab console, then Matlab will remember where GFtbox is, and on subsequent runs of Matlab you will not have to cd to the GFtbox directory before running it.
In addition to the tutorials given here, GFtbox comes with several example projects and the projects behind our published Figures. These projects appear on the Projects menu of the GFtbox window, in the Models submenu. There is a Help menu and when a new project is formed - the new associated interaction function is filled with comments that illustrate how the model can be written. In the GUI, mouseover text is provided for each of the GUI functions and the last item in every menu is a local help submenu. Documentation files can be found in the doc subdirectory of GFtbox.
Limitations of GFtbox
GFtbox is primarily for modelling curved finitely thick surfaces and does not handle bulk three-dimensional solids.
(More stuff to appear...)