Ready Reference Manual: Difference between revisions
No edit summary |
No edit summary |
||
| (53 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
'''''GFtbox''''' For modelling the growth of shapes. <br> | |||
[[GFtbox|'''''Details''''': what? How? Where?]]<br> | |||
[[GFtbox Tutorial pages|'''''Tutorials''''': from the beginning]]<br> | |||
[[GFtbox Example pages|'''''Examples''''': from publications]]<br> | |||
[https://sourceforge.net/p/gftbox/ '''''Download''''' from SourceForge]<br> | |||
=Types of morphogens= | |||
[[Types of morphogens and factors|Types of morphogens and factors click here]] | |||
=Summary of controls= | =Summary of controls= | ||
Roll the mouse over any control and information about the control will pop-up. | Roll the mouse over any control and information about the control will pop-up. | ||
==Models== | |||
* The larger the time step, the fewer iterations will be needed to simulate a given amount of time, but the less accurate the simulation will be. If the time step is too large, diffusion calculations may even become unstable. As a rule of thumb, the time step should be chosen small enough that: | |||
** The product of growth rate and time step is everywhere less than 0.1 -- that is, no part of the mesh grows by more than 10% in a single iteration. | |||
** In a single iteration, no part of the mesh rotates by more than 10 degrees. | |||
** The product of time step and the relative rate of change of any morphogen should be no more than 0.1. The rate of change may derive from the decay rate you have set, or from interactions programmed in the interaction function. | |||
** If ''k'' is the diffusion constant of a morphogen, ''D'' is the diameter of the mesh, and dt is the time step, ''k dt^2/D'' should be no more than 0.1. | |||
* Errors in the interaction function will cause the system to stop, and the interaction function will be disabled. The controls in the Interaction Function panel will all turn red to indicate this. | |||
** To re-enable the interaction function, click the Reset button at the top of the GUI. | |||
** To find the error | |||
*** There will be a message on the Matlab console saying in which line of the interaction function it detected an error. Open the interaction function in the Matlab editor, find that line, and set a breakpoint there or shortly before (by clicking in the left margin -- a red dot will appear). | |||
*** The next time the interaction function runs, Matlab will suspend execution at the breakpoint and go into debug mode. You can then examine the values of variables and step through the code one line at a time in order to discover why the error is happening. | |||
*** Matlab provides extensive tutorials on how to program and there is much information on the web. | |||
==Running models== | ==Running models== | ||
The normally'' green Run | The normally '''green Run pane'''l turns '''red''' whilst the model is running.<br> | ||
A '''yellow''' Run panel means that the model is running on a version of the GFtbox that is older than the model itself (look at the interaction header to see our source code version control number for the copy of GFtbox that created the model).<br> | A '''yellow''' Run panel means that the model is running on a version of the GFtbox that is older than the model itself (look at the interaction header to see our source code version control number for the copy of GFtbox that created the model).<br> | ||
An interaction function error (or any other error) will stop the model from running. It is then necessary to Reset using the''' Reset key - which will have turned red'''.<br><br> | An interaction function error (or any other error) will stop the model from running. It is then necessary to Reset using the''' Reset key - which will have turned red'''.<br><br> | ||
To run a model from the GUI | |||
*Step, execute one step of ''dt''. A whole lot of information pours out at the command console. Sometimes it takes a while to solve the equations. The process is monitored by a series of dots . . . . . . . Further information is provided below the Plotting panel, e.g. Step number, time per iteration, time per finite element, growth, etc. | *Step, execute one step of ''dt''. A whole lot of information pours out at the command console. Sometimes it takes a while to solve the equations. The process is monitored by a series of dots . . . . . . . Further information is provided below the Plotting panel, e.g. Step number, time per iteration, time per finite element, growth, etc. | ||
*Run for, and enter the number of steps into the box | *Run for, and enter the number of steps into the box. | ||
*Run until, and enter the model time to which you want to run | *Run until, and enter the model time to which you want to run. | ||
*Run to, and enter the multiple of the initial area you want to run to | *Run to, and enter the multiple of the initial area you want to run to. | ||
*Stop, stops execution at the end of the current step. | *Stop, stops execution at the end of the current step. | ||
*Call, makes a call to the interaction function but does NOT grow. This is useful for checking initial conditions - set patterns of morphogens. Having made a Call to the interaction function it is good practice to then Restart the model before running a model. | *Call, makes a call to the interaction function but does NOT grow. This is useful for checking initial conditions - set patterns of morphogens. Having made a Call to the interaction function it is good practice to then Restart the model before running a model. | ||
*Menu:Stages. '''We usually run models''' by setting up a set of, say 5, stages that we want stored. Then, having run the model we can revert back to any of the stages - make a change - and re-run starting from that point. The stages are stored in the project directory with the suffix *000# where # is the time. Stage times are stored with the project - and copied into the daughter project with Save As. | |||
**Compute more stages, pops up a box asking for a set of times that you want stored then starts computing | |||
**Request more stages, does the same but does not start computing. | |||
**Save experiment stages, saves all the current stage files in a subdirectory. | |||
**Import experiment stages, recovers a saved experiment. This pair of commands help document results and make figures. In addition, when the system runs on a cluster, the results are returned as experimental results to be accessed in this way. | |||
*'''What to do when the running system gets stuck'''. Usually this is because the shape is too regular and cannot initiate a bulge. | |||
**Interrupt processing using standard Matlab convention - Control-C. Then reset the ''GFtbox''. Panel:Select Tool:Mesh Editor and slightly modify the Z shape a random amount: click Random. | |||
==Outputs== | ==Outputs== | ||
===Plotting panel=== | ===Plotting panel=== | ||
To view | To view | ||
*Specified values, i.e. kapar or a particular morphogen (factor) select from drop down menu top right and tick Plot | *Specified values, i.e. kapar or a particular morphogen (factor) select from the drop down menu at the top right, and tick the Plot Current Factor checkbox. | ||
**To autoscale the axes, set the checkbox Panel:Plot Options:Auto axis range. To autozoom as well, click Menus:Plot:Auto zoom and centre. | |||
**To autoscale the morphogen colours. Panel:Plot Options:Auto color range. To fix the colour range the easiest is to click From Picture and unclick the Auto color range. Alternatively, unclick the Auto color range box and key in values you want - click elsewhere on the GUI to force a replot. | |||
**To find out the level of a morphogen at a particular point on the canvas. Select Tool:Simulation and select the morphogen of interest (top right pull down) and click on the point of interest. The value at that point will appear in bold just below the Panel:Timescale box. | |||
*Output variables, e.g. growth rate, tick Plot output value and make selection from the two drop down menus. | *Output variables, e.g. growth rate, tick Plot output value and make selection from the two drop down menus. | ||
*thickness, tick/untick Menu:Plot:Thickness. | |||
*thickness, tick/untick Menu:Plot:Thickness | |||
To show/hide | To show/hide | ||
*thickness, tick/untick Menu:Plot:Thickness | *thickness, tick/untick Menu:Plot:Thickness. | ||
*mesh, tick/untick FE edges | *mesh, tick/untick FE edges. | ||
*axes, tick/untick Menu:Plot:Show/Hide axes<br> | *axes, tick/untick Menu:Plot:Show/Hide axes.<br> | ||
*legend, tick/untick Menu:Plot:Show/Hide legend - also Set legend to choose heading<br> | *legend, tick/untick Menu:Plot:Show/Hide legend - also Set legend to choose heading.<br> | ||
To set/modify values of morphogens (best done in the interaction function) | To set/modify values of morphogens (best done in the interaction function) | ||
* Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on appropriate button (Add constant or Add radial, etc). | * Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on appropriate button (Add constant or Add radial, etc). | ||
* at individual nodes (vertices). Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on node.<br> | * at individual nodes (vertices). Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on node.<br> | ||
To clip the mesh (hide part of the mesh) | To clip the mesh (hide part of the mesh) | ||
*Tick Clipping plane and choose the plane with Az (azimuth), | *Tick Clipping plane and choose the plane with Az (azimuth), El (elevation), D (distance from origin). | ||
*Tick Clip and select the region to hide by using morphogen values (click on Mgens button) | *Tick Clip and select the region to hide by using morphogen values (click on Mgens button). | ||
===Snapshots=== | ===Snapshots=== | ||
Some video drivers do not support OpenGl in which case the screenshot can look wrong. The solution is to select GFtbox:'''Menus:Plot:Renderer:zbuffer''' (bottom of Plot menu).<br><br> | |||
Saved in png format in the project snapshots subdirectory | Saved in png format in the project snapshots subdirectory | ||
*Clicking Take snapshot makes a png file copy of the plotting screen. At the same time a copy of the interaction function (relabelled to have .txt as a suffix) is also made - to help document the snapshot. | *Clicking Take snapshot makes a png file copy of the plotting screen. At the same time a copy of the interaction function (relabelled to have .txt as a suffix) is also made - to help document the snapshot. | ||
*Publication quality screenshot is made by changing the action of the Snapshot button using Menu:Misc:Hi-res snapshots and choosing a number of dots per inch. | *Publication quality screenshot is made by changing the action of the Snapshot button using Menu:Misc:Hi-res snapshots and choosing a number of dots per inch. | ||
=== | ===Exporting meshes for external use=== | ||
The current mesh can be saved in various formats for use outside GFtbox, using the various Save... commands on the Mesh menu. | |||
*VRML, to | *VRML is a standard format for 3D graphics, for which there are free viewers (for example, [http://www.bitmanagement.com/en/products/interactive-3d-clients BS Contact] or ParallelGraphics' [http://www.cortona3d.com/Try-Buy/Downloads.aspx Cortona 3D Viewer]). It is also a format supported by [http://www.zcorp.com/en/home.aspx ZCorp]'s 3D printers. When you export to VRML, after providing a file name you will be presented with a dialog that allows you to rescale the object and also to change its thickness. Changing the thickness is only important for 3D printing, as a physical model will be too fragile if the mesh is too thin. | ||
*FIG | *OBJ is another standard format for 3D models, and can be imported into most 3D modelling software. | ||
*FIG is the Matlab format for figure files. These files can be reopened in Matlab. | |||
*MAT is Matlab's format for storing arbitrary Matlab objects. | |||
MAT is the only format that saves the entire mesh structure. The other formats save only its current graphical representation. | |||
===Movies=== | ===Movies=== | ||
By default movies are uncompressed | Some video drivers do not support OpenGl in which case the screenshot can look wrong. The solution is to select GFtbox:'''Menus:Plot:Renderer:zbuffer''' (bottom of Plot menu).<br><br> | ||
By default, movies are uncompressed, since this is the only format that is sure to play on all machines. However, uncompressed movies can easily be gigabytes in size. Various compressed formats can be selected in the Movie:Codec menu. Several formats are available, although not all of them are supported on all Matlab installations, and where supported, the resulting movies may not play in all movie-playing software. You only need to select a codec once, and it will apply to all subsequently created movies. | |||
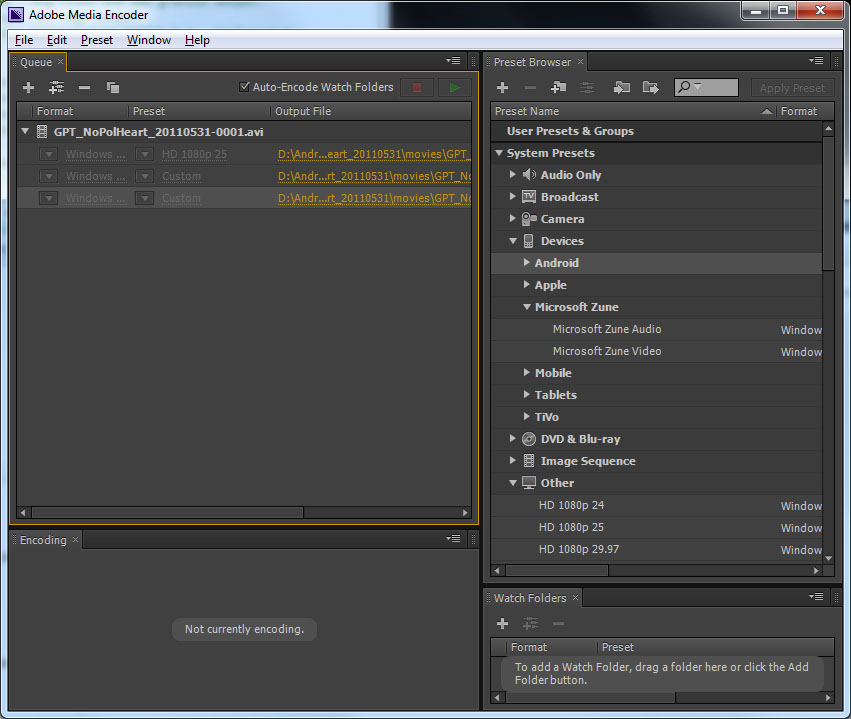
* | *For recent versions of Matlab (2010 and later) we<span style="color: Blue"> '''recommend the Motion JPEG AVI format'''</span>. (Older versions of Matlab do not provide it.). <span style="color: Blue">To convert .avi to .wmv (the preferred format for PowerPoint)</span> use Windows Movie Maker which is a free download from Microsoft, just drag the movie into the window and Save for a Computer (right hand end of control bar). Movie Maker allows limited editing of movies. Or use Adobe Media Encoder (converts anything to anything but definitely not free) which couples with Adobe Premier Pro to produce an incomprehensible professional editing suite. | ||
*Click Record Movie to start creating a movie. The button will change to Stop Movie. | **Adobe Media Encoder itself is simple. Recommended settings: | ||
{| border="0" width=100% | |||
|- | |||
|align="center"| | |||
|width="300" |*Adobe Media Encoder | |||
*Drag source file into left panel | |||
*Select output file format using the System Presets (right panel). | |||
**Either Microsoft Zune Video or | |||
**Other HD1080p 25. | |||
*Having selected a coder, Click on it in the Preset column of the Left panel and change the width/height settings to match the source movie (the details are in the summary at the top of the new window). You will probably have to unlink the aspect ratio button. | |||
*Finally, hit the green arrow top right of left panel. | |||
*You can set up multiple files to be converted at one time. | |||
*It will also output flash movies .flv. | |||
| | |||
|width="300pt"|[[Image:AdobeMediaEncoder.jpg|300px]] | |||
|} | |||
*Flash movie format cannot be created directly from Matlab, but there is an option to convert completed movies to Flash automatically, with the menu Movie:Convert to Flash. This option requires the ffmpeg program to be present on the Matlab command path. 'ffmpeg.exe' is available from the web.<br> | |||
Conversion from ''.avi'' to ''.flv'' is achieved with the help of | |||
MakeVideosAndFLV.m | |||
*Click Record Movie to start creating a movie. The current picture of the mesh will be the first frame of the movie. The button will change to Stop Movie. Every time a simulation step is performed, the picture of the mesh will be added to the movie file. Click Stop Movie to stop recording and close the movie file. The movie file will also be automatically closed if you close the project, load a new project, or close GFtbox. When the movie file is closed, the final frame will also be stored as a PNG image file, as well as a copy of the interaction function (with extension .txt instead of .m). | |||
*By default, the name of the movie file is chosen automatically, and consists of the project name followed by a numerical suffix. If you turn off the Auto-name checkbox below the Record Movie button, you will be prompted to given the movie a name. Movies (and the associated snapshots and interaction functions) are stored in a "movies" subdirectory of the project directory. | |||
<!-- | <!-- | ||
===Web and PowerPoint=== | |||
A copy of the interaction function suitable for inserting in a web page can be created by calling the function webify_interaction_function | |||
with embedded HTML | |||
Writing web and PowerPoint pages is facilitated by: | |||
*'''Output .avi for PowerPoint'''. Use Movie:Codec:Motion JPEG avi. This does not play properly on PowerPoint 2007 unless you run PowerPoint in 2003 compatibility mode. On the PC it can be converted to .wmf (the format that seems to be preferred by PowerPoint) using Windows Movie Maker. The movie is saved into the project movies subdirectory together with an image (.png) of the final frame and a copy of the interaction function (renamed .txt - to document the results). | |||
*Output '''flash (.flv) movies for web'''. Select Movie:Convert to Flash. This option requires the [http://www.ffmpeg.org/ ffmpeg] program to be on the current path. The movie is saved to the project directory together with an image (.png) of the first frame (for last frame see above). | |||
*Output''' web compatible copy of the interaction function''' use ''webify_interaction_function.m'' which outputs a txt file with comments marked up in HTML to appear in green and sections in blue. | |||
===Files=== | ===Files=== | ||
==Simulation== | ==Simulation== | ||
| Line 53: | Line 117: | ||
===Functions=== | ===Functions=== | ||
--> | --> | ||
=Useful interaction function programming constructs= | |||
The patterns of morphogens A and B are set up by <br> | [[Types of morphogens and factors|Types of morphogens and factors click here]]<br><br> | ||
{| border="0" cellpadding="1" cellspacing="1" | |||
|- valign="top" | |||
|width="150pt"|'''Pre-defined variables'''|| | |||
{| border="0" cellpadding="1" cellspacing="1" | |||
|- valign="top" | |||
|width="150pt"|dt|| computational step size | |||
|- | |||
|width="150pt"|realtime|| virtual time of the model | |||
|} | |||
|- valign="top" | |||
|width="150pt"|'''Pre-defined functions'''|| | |||
{| border="0" cellpadding="1" cellspacing="1" | |||
|- valign="top" | |||
|width="150pt"|Steps(m)|| current step number in Mesh m | |||
|- | |||
|width="150pt"|pro(k,id)|| promote by k in regions designated by factor, id (''pro(k,'''x''')=1+k'''x''''', i.e. promote by ''k=0'' and the value of ''pro()'' is ''1''). | |||
|- | |||
|width="150pt"|inh(k,id)|| inhibit by k in regions designated by factor, id (''inh(k,'''x''')=1/(1+k'''x''')'', i.e. inhibit by ''k=0'' and the value of ''inh()'' is ''1''). | |||
|- | |||
|width="150pt"|local_setproperties( m )|| initialise Mesh properties | |||
|- valign="top" | |||
|width="150pt"|leaf_*( m, ... )|| A large library of functions for manipulating the Mesh data structure, m. All of them are listed in the main Help menu of the GFtbox GUI, both in alphabetical order and categorised by topic. | |||
|} | |||
|} | |||
The patterns of morphogens A and B are often set up by using logical indexing, e.g.<br> | |||
id_a_p(m.nodes(:,1)<-0.03)=1; | id_a_p(m.nodes(:,1) < -0.03) = 1; | ||
id_b_p(m.nodes(:,2)<-0.01)=1; | id_b_p(m.nodes(:,2) < -0.01) = 1; | ||
where ''id_a_p'' is the A morphogen. ''m.node(:,1)'' refers to the ''x'' coordinates of all nodes (vertices) in the mesh<br> | where ''id_a_p'' is the vector of values of the A morphogen. ''m.node(:,1)'' refers to the ''x'' coordinates of all nodes (vertices) in the mesh.<br> | ||
The expression ''(m.nodes(:,1)<-0.03)'' means find all vertices with ''x'' coordinates that are less than -0.03.<br><br> | The expression ''(m.nodes(:,1) < -0.03)'' means find all vertices with ''x'' coordinates that are less than -0.03.<br><br> | ||
Similarly, ''(m.nodes(:,2)<-0.01)'' means find all vertices with ''y'' coordinates that are less than -0.01.<br><br> | Similarly, ''(m.nodes(:,2) < -0.01)'' means find all vertices with ''y'' coordinates that are less than -0.01.<br><br> | ||
And the pattern of polariser (P) is set up by<br> | And the pattern of polariser (P) is set up by<br> | ||
P((m.nodes(:,1)<-0.05)&(m.nodes(:,2)>0.03))=1; | P((m.nodes(:,1) < -0.05) & (m.nodes(:,2) > 0.03)) = 1; | ||
Where ''(m.nodes(:,1)<-0.05)&(m.nodes(:,2)>0.03)'' means find all vertices with | Where ''(m.nodes(:,1) < -0.05) & (m.nodes(:,2) > 0.03)'' means find all vertices with x coordinate less than -0.05 and y coordinate greater than 0.03.<br><br> | ||
Thus the full code describing the model is: | Thus the full code describing the model is: | ||
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. | if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. | ||
id_a_p(m.nodes(:,1)<-0.03)=1; % setup region for A where identity factor A is represented by id_a_p | id_a_p(m.nodes(:,1) < -0.03) = 1; % setup region for A where identity factor A is represented by id_a_p | ||
id_b_p(m.nodes(:,2)<-0.01)=1; % setup region for B | id_b_p(m.nodes(:,2) < -0.01) = 1; % setup region for B | ||
else | else | ||
% @@KRN Growth Regulatory Network | % @@KRN Growth Regulatory Network | ||
| Line 78: | Line 168: | ||
knor_p(:) = 0; % thickness not growing | knor_p(:) = 0; % thickness not growing | ||
end | end | ||
=Default '''''GFtbox''''' parameters= | |||
==Functions== | |||
function leaf_setproperty | |||
m = leaf_setproperty( m, ... ) | |||
Set global properties of the leaf. | |||
The arguments are a series of name/value pairs. | |||
The property names that this applies to are: | |||
'poisson' Poisson's ratio. The normal value is 0.35 and | |||
there is little need to change this. | |||
'bulkmodulus' The bulk modulus. The normal value is 3000 and | |||
there is little need to change this. | |||
'validate' Whether to validate the mesh after every iteration. | |||
This is for debugging purposes and should normally | |||
be off. | |||
'displayedgrowth' Specifies which morphogen to plot. | |||
...and many others I have omitted to document. | |||
Example: | |||
m = leaf_setproperty( m, 'poisson', 0.49 ); | |||
Equivalent GUI operation: several GUI elements are implemented by this | |||
command: | |||
poisson: Text box in "Mesh editor" panel. | |||
bulkmodulus: No GUI equivalent. | |||
residstrain: "Strain retention" in "Simulation" panel. | |||
validate: No GUI equivalent. | |||
displayedgrowth: "Displayed m'gen" menu in "Morphogens" panel. | |||
...etc. | |||
Topics: Misc. | |||
and | |||
leaf_getproperty | |||
s = leaf_getproperty( m, ... ) | |||
Read various properties of the mesh. | |||
The arguments are any property names that can be passed to | |||
leaf_setproperty. The result is a structure whose fields | |||
are those names and whose values are the values they have in m. | |||
Unrecognised names will be ignored. | |||
Unlike most leaf_* commands, this command does not modify m and | |||
therefore does not return a new value of m. | |||
Topics: Misc. | |||
All the main properties can be seen by looking at ''definePropertyLists.m'', e.g. | |||
function definePropertyLists() | |||
%definePropertyLists() | |||
% This defines some tables of mesh properties, mainly gGlobalProps (the | |||
% default value of m.globalProps), gGlobalDynamicProps (the default value | |||
% of m.globalDynamicProps), and gDefaultPlotOptions (the default value of | |||
% m.plotdefaults). Documentation is interspersed with the definitions | |||
% which can be automatically extracted and used to populate a Help menu. | |||
global gGlobalProps gGlobalDynamicProps gDefaultPlotOptions | |||
global gOurViewParams gMatlabViewParams | |||
global gOurViewParamNames gMatlabViewParamNames | |||
global gGAUSS_INFO; | |||
gGAUSS_INFO = computeGaussInfo(); | |||
secondlayercolors = [[0.1,1,0.1];[1,0.1,0.1]]; | |||
secondlayercolorvariation = 0.05; | |||
gGlobalProps = struct( ... % ITEM leaf_setproperties | |||
'trinodesvalid', false, ... % DYNAMIC, UNSAVED | |||
'prismnodesvalid', false, ... % DYNAMIC, UNSAVED | |||
'thicknessRelative', 0.5, ... | |||
'thicknessArea', 0, ... | |||
'thicknessMode', 'physical', ... | |||
'activeGrowth', 1, ... | |||
'displayedGrowth', 1, ... | |||
'displayedMulti', [], ... | |||
'allowNegativeGrowth', true, ... | |||
... % If true, the growth morphogens KAPAR, KAPER, KBPAR, and KBPER | |||
... % can be negative. If false, any negative values arising in the | |||
... % computation are replaced by zero. | |||
'usePrevDispAsEstimate', true, ... | |||
... % If true, the computation of elastic deformation will use the | |||
... % displacements computed on the previous timestep as the initial | |||
... % estimate for the current timestep. If false, the initial estimate | |||
... % will be zero. | |||
'perturbInitGrowthEstimate', 0.00001, ... | |||
... % This defines the amount of random perturbation added to the initial | |||
... % estimate for the computation of elastic deformation, as an absolute amount. | |||
'perturbRelGrowthEstimate', 0.01, ... | |||
... % This defines the amount of random perturbation added to the initial | |||
... % estimate for the computation of elastic deformation, as a relative amount. | |||
'perturbDiffusionEstimate', 0.0001, ... | |||
... % This defines the amount of random perturbation added to the initial | |||
... % estimate for the computation of diffusion. | |||
'resetRand', false, ... | |||
... % If true, the random number generator will be reset to a fixed initial | |||
... % state before each use made of it. This allows exact reproducibility | |||
... % of a simulation. If false, it is never reset. | |||
'mingradient', 0, ... | |||
'relativepolgrad', false, ... | |||
'usefrozengradient', true, ... | |||
'userpolarisation', false, ... | |||
'thresholdsq', 0, ... | |||
'splitmargin', 1.4, ... | |||
... % When splitting of long edges is enabled, an edge will be bisected | |||
... % if its length is this amount times the threshold. The default | |||
... % value is approximately sqrt(2), so that after splitting the two new | |||
... % edges are below the threshold by the same ratio. | |||
'splitmorphogen', '', ... | |||
... % The value of this is the name of a morphogen to be used to influence | |||
... % edge-splitting. Every edge where this morphogen exceeds a specified | |||
... % theshold at both ends will be split. See also 'thresholdmgen'. | |||
'thresholdmgen', 0.5, ... | |||
... % This is the threshold value for morphogen-specified edge-splitting. | |||
... % See also 'splitmorphogen'. | |||
'bulkmodulus', 1, ... | |||
... % IGNORE | |||
'unitbulkmodulus', true, ... | |||
... % IGNORE | |||
'poissonsRatio', 0.3, ... | |||
... % This sets Poisson's ratio for the material, a constant value everywhere. | |||
'starttime', 0, ... | |||
... % This defines the time at the start of the simulation. | |||
'timestep', 0.01, ... | |||
... % This is the time step of the simulation. It should be chosen small enough | |||
... % that in a single time step, no part of the mesh grows by more than 10% in | |||
... % any direction, and rotates by no more than 10 degrees. For more accurate | |||
... % runs, these criteriashould be reduced to 5% and 5 degrees. | |||
'timeunitname', '', ... | |||
... % This is the name of the unit of time, and should be given in the singular. | |||
'distunitname', 'mm', ... | |||
... % This is the name of the unit of length. | |||
'scalebarvalue', 0, ... | |||
'validateMesh', true, ... | |||
... % This is for debugging only. If true, a large set of validation checks | |||
... % is run after each simulation step, to verify the integrity of the data | |||
... % structures. | |||
'rectifyverticals', false, ... | |||
... % If true, after each simulation step the pairs of vertexes on opposite | |||
... % side of the mesh are moved as necessary to ensure that the line joinging | |||
... % them is perpendicular to the mesh. The midpoint of this line does not move. | |||
... % This should be thought of as revising the decomposition of the material | |||
... % into finite elements, not as a deformation of the material. | |||
... % WARNING | |||
... % BEFORE 2013 this was always treated as true (even if set false). | |||
... % From 2013 onwards it is set true by default. | |||
... % | |||
'allowSplitLongFEM', true, ... | |||
... % This boolean flag specifies whether edges exceeeding a certain length | |||
... % threshold should be split after each simulation step. | |||
'longSplitThresholdPower', 0, ... | |||
'allowSplitBentFEM', false, ... | |||
'allowSplitBio', true, ... | |||
'allowFlipEdges', false, ... | |||
'allowElideEdges', true, ... | |||
'mincellangle', 0.2, ... | |||
'alwaysFlat', false, ... | |||
'flattenforceconvex', true, ... | |||
'flatten', false, ... % IGNORE. Obsolete but harmless | |||
'flattenratio', 1, ... % IGNORE. Obsolete but harmless | |||
'useGrowthTensors', false, ... | |||
'plasticGrowth', false, ... | |||
... % If true, the material is modelled as a fluid flowing with Reynolds | |||
... % number equal to zero, which is the limiting behaviour as Poisson's | |||
... % ratio tends to 0.5. The default is to model it as an elastic solid. | |||
'totalinternalrotation', 0, ... % IGNORE | |||
'stepinternalrotation', 2, ... % IGNORE radians | |||
'showinternalrotation', false, ... % IGNORE | |||
'performinternalrotation', false, ... % IGNORE | |||
'internallyrotated', false, ... % IGNORE | |||
'maxFEcells', int32(0), ... | |||
... % This specifies the maximum number of finite elements the mesh is | |||
... % allowed to contain. When it reaches this number, no more edge-splitting | |||
... % will be done. A value of zero means the number is unlimited. | |||
'inittotalcells', int32(0), ... | |||
'bioApresplitproc', '', ... | |||
... % This should be the name of a function which will be called immediately | |||
... % before determining whether any biological cells should be split. This | |||
... % function can be used to override the default cell-splitting method. The | |||
... % method of writing this function is rather complex and not documented here. | |||
'bioApostsplitproc', '', ... | |||
... % This should be the name of a function which will be called immediately | |||
... % after having split some biological cells. This | |||
... % function can be used to override the default cell-splitting method. The | |||
... % method of writing this function is rather complex and not documented here. | |||
'maxBioAcells', int32(0), ... | |||
... % This specifies the maximum number of biological cells the mesh is | |||
... % allowed to contain. When it reaches this number, no more cell-splitting | |||
... % will be done. A value of zero means the number is unlimited. | |||
'colors', secondlayercolors, ... | |||
'colorvariation', secondlayercolorvariation, ... | |||
'colorparams', makesecondlayercolorparams( secondlayercolors, secondlayercolorvariation ), ... | |||
'biocolormode', 'auto', ... | |||
'freezing', 0, ... | |||
'canceldrift', false, ... | |||
... % If true, after each simulation step, the mesh is repositioned if necessary | |||
... % to force its centre (defined as the average of all of its vertexes) to | |||
... % remain stationary. | |||
'mgen_interaction', '', ... | |||
... % READ-ONLY. This is a handle to the interaction function, and should not be | |||
... % modified by the user. | |||
'mgen_interactionName', '', ... | |||
... % READ-ONLY. This is the name of the interaction function, and should not be | |||
... % modified by the user. | |||
'allowInteraction', true, ... | |||
... % This specifies whether the interaction function should be called during each | |||
... % simulation step. | |||
'interactionValid', true, ... % UNSAVED | |||
... % READ-ONLY. If an error is detected in the interaction function, this | |||
... % flag will be set to false. It reverts to true of GFtbox detects that | |||
... % the user has edited it, or when the Reset button in the GUI is clicked. | |||
'gaussInfo', gGAUSS_INFO, ... % IGNORE | |||
'stitchDFs', [], ... | |||
'D', zeros(6,6), ... % IGNORE | |||
'C', zeros(6,6), ... % IGNORE | |||
'G', zeros(6,1), ... % IGNORE | |||
'solver', 'cgs', ... | |||
... % This specifies which solver to user when computing elastic deformation. | |||
'solverprecision', 'double', ... | |||
'solvertolerance', 0.001, ... | |||
'solvertolerancemethod', 'max', ... | |||
'diffusiontolerance', 0.00001, ... | |||
'allowsparse', true, ... | |||
'maxIters', int32(0), ... | |||
'maxsolvetime', 1000, ... | |||
'cgiters', int32(0), ... | |||
'simsteps', int32(0), ... | |||
'stepsperrender', int32(0), ... | |||
'growthEnabled', true, ... | |||
... % This specifies whether elastic growth is to be computed during a | |||
... % simulation step. | |||
'diffusionEnabled', true, ... | |||
... % This specifies whether diffusion is to be computed during a | |||
... % simulation step. | |||
'flashmovie', false, ... | |||
... % This specifies whether all movies should be automatically | |||
... % converted to Flash format on completion. | |||
'makemovie', false, ... | |||
... % This specifies whether a frame is to be saved to a movie file | |||
... % after each simulation step. | |||
'moviefile', '', ... | |||
... % The name of the current movie file (empty when a movie is not being recorded). | |||
'codec', 'None', ... | |||
... % The codec to be uesd when recording a movie file. | |||
'autonamemovie', true, ... | |||
... % If true, when a movie is started, a name will be automatically | |||
... % generated for the movie file. If false, the user will be presented | |||
... % with a file dialog to choose the name. | |||
'overwritemovie', false, ... | |||
... % When this is true, no check will be made to see if an automatically | |||
... % generated movie file name already exists. If false, the name will be | |||
... % modified by appending a unique number in order to prevent any existing | |||
... % fie from being overwritten. | |||
'framesize', [], ... % IGNORE UNSAVED | |||
'mov', [], ... % IGNORE UNSAVED | |||
'boingNeeded', false, ... % IGNORE | |||
'initialArea', 0, ... | |||
'bendunitlength', 0, ... | |||
'targetRelArea', 1, ... % UNSAVED | |||
'defaultinterp', 'min', ... | |||
'readonly', false, ... | |||
'projectdir', '', ... | |||
'modelname', '', ... | |||
'allowsave', true, ... % IGNORE UNSAVED | |||
'addedToPath', false, ... % IGNORE UNSAVED | |||
'bendsplit', 0.3, ... | |||
'usepolfreezebc', false, ... | |||
'dorsaltop', true, ... | |||
'defaultazimuth', -45, ... | |||
'defaultelevation', 33.75, ... | |||
'defaultroll', 0, ... | |||
'defaultViewParams', gMatlabViewParams, ... | |||
'comment', '', ... | |||
'legendTemplate', '%T: %q\n%m', ... | |||
... % This is the format string used to define the legend appearing | |||
... % at the top of the picture area. | |||
'bioAsplitcells', true, ... | |||
... % Allow splitting of biological cells. | |||
'bioApullin', 4/28, ... | |||
'bioAfakepull', 0.7/(2*sqrt(3)), ... | |||
'interactive', false, ... | |||
... % READ-ONLY. Says whether GFtbox is running interactively. Obsolete? | |||
'coderevision', int32(0), ... | |||
... % READ-ONLY. The current revision number of GFtbox. | |||
'coderevisiondate', '', ... | |||
... % READ-ONLY. The current revision date of GFtbox. | |||
'modelrevision', int32(0), ... | |||
... % READ-ONLY. The latest revision of GFtbox that this model was | |||
... % operated on with. Backward compatibility is retained: old models | |||
... % should always be loadable into new versions of GFtbox. The reverse | |||
... % is not necessarily the case. This field will tell you if the model | |||
... % has been edited with a newer version of GFtbox that you are currently | |||
... % running. | |||
'modelrevisiondate', '', ... | |||
... % READ-ONLY. The date of the latest revision of GFtbox that this model was | |||
... % operated on with. See also 'modelrevision'. | |||
'savedrunname', '', ... % IGNORE | |||
'savedrundesc', '' ... % IGNORE | |||
); | |||
gGlobalProps.vxgrad(:,:,1) = gradN( [ 0; 0; 1 ] ); | |||
gGlobalProps.vxgrad(:,:,2) = gradN( [ 1; 0; 1 ] ); | |||
gGlobalProps.vxgrad(:,:,3) = gradN( [ 0; 1; 1 ] ); | |||
gGlobalProps.vxgrad(:,:,4) = gradN( [ 0; 0; -1 ] ); | |||
gGlobalProps.vxgrad(:,:,5) = gradN( [ 1; 0; -1 ] ); | |||
gGlobalProps.vxgrad(:,:,6) = gradN( [ 0; 1; -1 ] ); | |||
gGlobalDynamicProps = struct( ... % ITEM leaf_setproperties | |||
'currenttime', 0.0, ... % DYNAMIC | |||
'currentIter', 0, ... % DYNAMIC | |||
'laststagesuffix', '', ... % DYNAMIC, UNSAVED | |||
'currentArea', 0, ... % DYNAMIC | |||
'previousArea', 0, ... % DYNAMIC | |||
'thicknessAbsolute', 0, ... % DYNAMIC? | |||
'cellscale', 0, ... % DYNAMIC | |||
'locatenode', 0, ... % DYNAMIC | |||
'locateDFs', [], ... % DYNAMIC | |||
'staticreadonly', false, ... % DYNAMIC | |||
'commandok', true, ... % DYNAMIC | |||
'doinit', true ... % DYNAMIC | |||
); | |||
gOurViewParams = struct( ... | |||
'azimuth', -45, ... | |||
'elevation', 33.75, ... | |||
'roll', 0, ... | |||
'fov', 10, ... | |||
'pan', [0 0], ... | |||
'targetdistance', 0, ... | |||
'camdistance', 4*sqrt(3)/(2*tan(5*pi/180)), ... | |||
'projection', 'orthographic' ); | |||
gMatlabViewParams = cameraParamsFromOurViewParams( gOurViewParams ); | |||
gOurViewParamNames = fieldnames( gOurViewParams ); | |||
gMatlabViewParamNames = fieldnames( gMatlabViewParams ); | |||
gDefaultPlotOptions = struct( ... % ITEM leaf_plotoptions | |||
'drawtensoraxes', false, ... | |||
'unitcrosses', true, ... | |||
... % 'tensorquantity', 'resultantgrowthrate', ... % New 2009-11-11 | |||
... % 'tensorproperty', 'total', ... | |||
'outputquantity', 'resultantgrowthrate', ... | |||
'outputaxes', 'total', ... | |||
'morphogen', 1, ... | |||
... % 'rawstuff', [], ... % Obsolete. | |||
'drawleaf', 1, ... | |||
'drawedges', 1, ... | |||
'drawseams', 1, ... | |||
'fillleaf', 1, ... | |||
'drawgradients', 0, ... | |||
'unitgradients', 0, ... | |||
'multibrighten', 0.1, ... | |||
... % 'monochrome', true, ... | |||
'drawcolorbar', true, ... | |||
'monocolors', [[0 0 1];[1 0 0]], ... | |||
'canvascolor', [1 1 1], ... | |||
'cmap', [], ... | |||
'crange', [], ... | |||
'zerowhite', 0, ... | |||
'cmaptype', 'monochrome', ... | |||
'sparsedistance', 0, ... | |||
'staticdecor', true, ... | |||
'axisRange', [], ... | |||
'axisVisible', 1, ... | |||
'alpha', 0.8, ... | |||
'ambientstrength', 0.8, ... | |||
'azimuth', -45, ... | |||
'elevation', 33.75, ... | |||
'roll', 0, ... | |||
'ourViewParams', gOurViewParams, ... | |||
'matlabViewParams', gMatlabViewParams, ... | |||
'autozoom', false, ... | |||
'autocentre', false, ... | |||
'autoScale', 1, ... | |||
'autoColorRange', 1, ... | |||
'drawsecondlayer', 0, ... | |||
'cellsonbothsides', true, ... | |||
'layeroffset', 0.2, ... | |||
'FEthicklinesize', 2, ... | |||
'FEthinlinesize', 1, ... | |||
'FElinecolor', [0 0 0], ... | |||
'seamlinesize', 5, ... | |||
'seamlinecolor', [1 0 0], ... | |||
'bioAlinesize', 1, ... | |||
'bioAlinecolor', [0.2 0.2 0.2], ... | |||
'bioAnewlinecolor', [0.2 0.2 0.2], ... | |||
'bioApointsize', 3, ... | |||
'bioApointcolor', [0.2 0.2 0.2], ... | |||
'bioAnewpointcolor', [0.2 0.2 0.2], ... | |||
'bioAalpha', 1, ... | |||
'axescolor', [0 0 0.3], ... | |||
'decorateAside', false, ... | |||
'drawnormals', 0, ... | |||
'drawmutant', 1, ... | |||
'drawdisplacements', 0, ... | |||
'drawlegend', 1, ... | |||
'drawscalebar', 1, ... | |||
'thick', 1, ... | |||
'nodenumbering', 0, ... | |||
'nodenumbercolor', [1 0 0], ... | |||
'edgenumbering', 0, ... | |||
'edgenumbercolor', [0 0.4 0.1], ... | |||
'FEnumbering', 0, ... | |||
'FEnumbercolor', [0 0 0.4], ... | |||
'texture', 0, ... | |||
'bgcolor', [1 1 1], ... | |||
'emptycolor', [0.7, 0.8, 1], ... | |||
'clippingAzimuth', 0, ... | |||
'clippingElevation', 0, ... | |||
'clippingDistance', -0.01, ... | |||
'clipbymgen', false, ... | |||
'clipmgens', [], ... | |||
'clipmgenthreshold', 0, ... | |||
'clipmgenabove', true, ... | |||
'clipmgenall', true, ... | |||
'doclip', false, ... | |||
'uicontrols', true, ... | |||
'light', false, ... | |||
'lightmode', 'gouraud', ... | |||
'invisibleplot', false, ... | |||
'decorscale', 1, ... | |||
'highlightthickness', 3, ... | |||
'arrowthickness', 2, ... | |||
'crossthickness', 1, ... | |||
'arrowheadsize', 0.5, ... | |||
'arrowheadratio', 0.3, ... | |||
'highgradcolor', [0 0 0.3], ... | |||
'lowgradcolor', [0.5 0 0], ... | |||
'hiresdpi', 400, ... | |||
'mgenownsideonly', 'false', ... | |||
'taper', 'true', ... | |||
'anisotropythreshold', 0, ... | |||
'linesmoothing', 'on', ... | |||
'userplotproc', [] ); | |||
end | |||
Latest revision as of 14:30, 20 March 2013
GFtbox For modelling the growth of shapes.
Details: what? How? Where?
Tutorials: from the beginning
Examples: from publications
Download from SourceForge
Types of morphogens
Types of morphogens and factors click here
Summary of controls
Roll the mouse over any control and information about the control will pop-up.
Models
- The larger the time step, the fewer iterations will be needed to simulate a given amount of time, but the less accurate the simulation will be. If the time step is too large, diffusion calculations may even become unstable. As a rule of thumb, the time step should be chosen small enough that:
- The product of growth rate and time step is everywhere less than 0.1 -- that is, no part of the mesh grows by more than 10% in a single iteration.
- In a single iteration, no part of the mesh rotates by more than 10 degrees.
- The product of time step and the relative rate of change of any morphogen should be no more than 0.1. The rate of change may derive from the decay rate you have set, or from interactions programmed in the interaction function.
- If k is the diffusion constant of a morphogen, D is the diameter of the mesh, and dt is the time step, k dt^2/D should be no more than 0.1.
- Errors in the interaction function will cause the system to stop, and the interaction function will be disabled. The controls in the Interaction Function panel will all turn red to indicate this.
- To re-enable the interaction function, click the Reset button at the top of the GUI.
- To find the error
- There will be a message on the Matlab console saying in which line of the interaction function it detected an error. Open the interaction function in the Matlab editor, find that line, and set a breakpoint there or shortly before (by clicking in the left margin -- a red dot will appear).
- The next time the interaction function runs, Matlab will suspend execution at the breakpoint and go into debug mode. You can then examine the values of variables and step through the code one line at a time in order to discover why the error is happening.
- Matlab provides extensive tutorials on how to program and there is much information on the web.
Running models
The normally green Run panel turns red whilst the model is running.
A yellow Run panel means that the model is running on a version of the GFtbox that is older than the model itself (look at the interaction header to see our source code version control number for the copy of GFtbox that created the model).
An interaction function error (or any other error) will stop the model from running. It is then necessary to Reset using the Reset key - which will have turned red.
To run a model from the GUI
- Step, execute one step of dt. A whole lot of information pours out at the command console. Sometimes it takes a while to solve the equations. The process is monitored by a series of dots . . . . . . . Further information is provided below the Plotting panel, e.g. Step number, time per iteration, time per finite element, growth, etc.
- Run for, and enter the number of steps into the box.
- Run until, and enter the model time to which you want to run.
- Run to, and enter the multiple of the initial area you want to run to.
- Stop, stops execution at the end of the current step.
- Call, makes a call to the interaction function but does NOT grow. This is useful for checking initial conditions - set patterns of morphogens. Having made a Call to the interaction function it is good practice to then Restart the model before running a model.
- Menu:Stages. We usually run models by setting up a set of, say 5, stages that we want stored. Then, having run the model we can revert back to any of the stages - make a change - and re-run starting from that point. The stages are stored in the project directory with the suffix *000# where # is the time. Stage times are stored with the project - and copied into the daughter project with Save As.
- Compute more stages, pops up a box asking for a set of times that you want stored then starts computing
- Request more stages, does the same but does not start computing.
- Save experiment stages, saves all the current stage files in a subdirectory.
- Import experiment stages, recovers a saved experiment. This pair of commands help document results and make figures. In addition, when the system runs on a cluster, the results are returned as experimental results to be accessed in this way.
- What to do when the running system gets stuck. Usually this is because the shape is too regular and cannot initiate a bulge.
- Interrupt processing using standard Matlab convention - Control-C. Then reset the GFtbox. Panel:Select Tool:Mesh Editor and slightly modify the Z shape a random amount: click Random.
Outputs
Plotting panel
To view
- Specified values, i.e. kapar or a particular morphogen (factor) select from the drop down menu at the top right, and tick the Plot Current Factor checkbox.
- To autoscale the axes, set the checkbox Panel:Plot Options:Auto axis range. To autozoom as well, click Menus:Plot:Auto zoom and centre.
- To autoscale the morphogen colours. Panel:Plot Options:Auto color range. To fix the colour range the easiest is to click From Picture and unclick the Auto color range. Alternatively, unclick the Auto color range box and key in values you want - click elsewhere on the GUI to force a replot.
- To find out the level of a morphogen at a particular point on the canvas. Select Tool:Simulation and select the morphogen of interest (top right pull down) and click on the point of interest. The value at that point will appear in bold just below the Panel:Timescale box.
- Output variables, e.g. growth rate, tick Plot output value and make selection from the two drop down menus.
- thickness, tick/untick Menu:Plot:Thickness.
To show/hide
- thickness, tick/untick Menu:Plot:Thickness.
- mesh, tick/untick FE edges.
- axes, tick/untick Menu:Plot:Show/Hide axes.
- legend, tick/untick Menu:Plot:Show/Hide legend - also Set legend to choose heading.
To set/modify values of morphogens (best done in the interaction function)
- Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on appropriate button (Add constant or Add radial, etc).
- at individual nodes (vertices). Choose morphogen (list box), Click on Select Tool:Factors, '---' list box to select action and click on node.
To clip the mesh (hide part of the mesh)
- Tick Clipping plane and choose the plane with Az (azimuth), El (elevation), D (distance from origin).
- Tick Clip and select the region to hide by using morphogen values (click on Mgens button).
Snapshots
Some video drivers do not support OpenGl in which case the screenshot can look wrong. The solution is to select GFtbox:Menus:Plot:Renderer:zbuffer (bottom of Plot menu).
Saved in png format in the project snapshots subdirectory
- Clicking Take snapshot makes a png file copy of the plotting screen. At the same time a copy of the interaction function (relabelled to have .txt as a suffix) is also made - to help document the snapshot.
- Publication quality screenshot is made by changing the action of the Snapshot button using Menu:Misc:Hi-res snapshots and choosing a number of dots per inch.
Exporting meshes for external use
The current mesh can be saved in various formats for use outside GFtbox, using the various Save... commands on the Mesh menu.
- VRML is a standard format for 3D graphics, for which there are free viewers (for example, BS Contact or ParallelGraphics' Cortona 3D Viewer). It is also a format supported by ZCorp's 3D printers. When you export to VRML, after providing a file name you will be presented with a dialog that allows you to rescale the object and also to change its thickness. Changing the thickness is only important for 3D printing, as a physical model will be too fragile if the mesh is too thin.
- OBJ is another standard format for 3D models, and can be imported into most 3D modelling software.
- FIG is the Matlab format for figure files. These files can be reopened in Matlab.
- MAT is Matlab's format for storing arbitrary Matlab objects.
MAT is the only format that saves the entire mesh structure. The other formats save only its current graphical representation.
Movies
Some video drivers do not support OpenGl in which case the screenshot can look wrong. The solution is to select GFtbox:Menus:Plot:Renderer:zbuffer (bottom of Plot menu).
By default, movies are uncompressed, since this is the only format that is sure to play on all machines. However, uncompressed movies can easily be gigabytes in size. Various compressed formats can be selected in the Movie:Codec menu. Several formats are available, although not all of them are supported on all Matlab installations, and where supported, the resulting movies may not play in all movie-playing software. You only need to select a codec once, and it will apply to all subsequently created movies.
- For recent versions of Matlab (2010 and later) we recommend the Motion JPEG AVI format. (Older versions of Matlab do not provide it.). To convert .avi to .wmv (the preferred format for PowerPoint) use Windows Movie Maker which is a free download from Microsoft, just drag the movie into the window and Save for a Computer (right hand end of control bar). Movie Maker allows limited editing of movies. Or use Adobe Media Encoder (converts anything to anything but definitely not free) which couples with Adobe Premier Pro to produce an incomprehensible professional editing suite.
- Adobe Media Encoder itself is simple. Recommended settings:
- Flash movie format cannot be created directly from Matlab, but there is an option to convert completed movies to Flash automatically, with the menu Movie:Convert to Flash. This option requires the ffmpeg program to be present on the Matlab command path. 'ffmpeg.exe' is available from the web.
Conversion from .avi to .flv is achieved with the help of
MakeVideosAndFLV.m
- Click Record Movie to start creating a movie. The current picture of the mesh will be the first frame of the movie. The button will change to Stop Movie. Every time a simulation step is performed, the picture of the mesh will be added to the movie file. Click Stop Movie to stop recording and close the movie file. The movie file will also be automatically closed if you close the project, load a new project, or close GFtbox. When the movie file is closed, the final frame will also be stored as a PNG image file, as well as a copy of the interaction function (with extension .txt instead of .m).
- By default, the name of the movie file is chosen automatically, and consists of the project name followed by a numerical suffix. If you turn off the Auto-name checkbox below the Record Movie button, you will be prompted to given the movie a name. Movies (and the associated snapshots and interaction functions) are stored in a "movies" subdirectory of the project directory.
Useful interaction function programming constructs
Types of morphogens and factors click here
| Pre-defined variables |
| ||||||||||
| Pre-defined functions |
|
The patterns of morphogens A and B are often set up by using logical indexing, e.g.
id_a_p(m.nodes(:,1) < -0.03) = 1;
id_b_p(m.nodes(:,2) < -0.01) = 1;
where id_a_p is the vector of values of the A morphogen. m.node(:,1) refers to the x coordinates of all nodes (vertices) in the mesh.
The expression (m.nodes(:,1) < -0.03) means find all vertices with x coordinates that are less than -0.03.
Similarly, (m.nodes(:,2) < -0.01) means find all vertices with y coordinates that are less than -0.01.
And the pattern of polariser (P) is set up by
P((m.nodes(:,1) < -0.05) & (m.nodes(:,2) > 0.03)) = 1;
Where (m.nodes(:,1) < -0.05) & (m.nodes(:,2) > 0.03) means find all vertices with x coordinate less than -0.05 and y coordinate greater than 0.03.
Thus the full code describing the model is:
if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code.
id_a_p(m.nodes(:,1) < -0.03) = 1; % setup region for A where identity factor A is represented by id_a_p
id_b_p(m.nodes(:,2) < -0.01) = 1; % setup region for B
else
% @@KRN Growth Regulatory Network
kapar_p(:) = id_a_l .* inh(1,id_b_l); % growth rate
kaper_p(:) = kapar_p; % isotropic growth
kbpar_p(:) = kapar_p; % same on both sides of the sheet
kbper_p(:) = kapar_p; % same
knor_p(:) = 0; % thickness not growing
end
Default GFtbox parameters
Functions
function leaf_setproperty
m = leaf_setproperty( m, ... )
Set global properties of the leaf.
The arguments are a series of name/value pairs.
The property names that this applies to are:
'poisson' Poisson's ratio. The normal value is 0.35 and
there is little need to change this.
'bulkmodulus' The bulk modulus. The normal value is 3000 and
there is little need to change this.
'validate' Whether to validate the mesh after every iteration.
This is for debugging purposes and should normally
be off.
'displayedgrowth' Specifies which morphogen to plot.
...and many others I have omitted to document.
Example:
m = leaf_setproperty( m, 'poisson', 0.49 );
Equivalent GUI operation: several GUI elements are implemented by this
command:
poisson: Text box in "Mesh editor" panel.
bulkmodulus: No GUI equivalent.
residstrain: "Strain retention" in "Simulation" panel.
validate: No GUI equivalent.
displayedgrowth: "Displayed m'gen" menu in "Morphogens" panel.
...etc.
Topics: Misc.
and leaf_getproperty
s = leaf_getproperty( m, ... ) Read various properties of the mesh. The arguments are any property names that can be passed to leaf_setproperty. The result is a structure whose fields are those names and whose values are the values they have in m. Unrecognised names will be ignored. Unlike most leaf_* commands, this command does not modify m and therefore does not return a new value of m. Topics: Misc.
All the main properties can be seen by looking at definePropertyLists.m, e.g.
function definePropertyLists() %definePropertyLists() % This defines some tables of mesh properties, mainly gGlobalProps (the % default value of m.globalProps), gGlobalDynamicProps (the default value % of m.globalDynamicProps), and gDefaultPlotOptions (the default value of % m.plotdefaults). Documentation is interspersed with the definitions % which can be automatically extracted and used to populate a Help menu.
global gGlobalProps gGlobalDynamicProps gDefaultPlotOptions global gOurViewParams gMatlabViewParams global gOurViewParamNames gMatlabViewParamNames global gGAUSS_INFO;
gGAUSS_INFO = computeGaussInfo();
secondlayercolors = [[0.1,1,0.1];[1,0.1,0.1]]; secondlayercolorvariation = 0.05;
gGlobalProps = struct( ... % ITEM leaf_setproperties
'trinodesvalid', false, ... % DYNAMIC, UNSAVED
'prismnodesvalid', false, ... % DYNAMIC, UNSAVED
'thicknessRelative', 0.5, ...
'thicknessArea', 0, ...
'thicknessMode', 'physical', ...
'activeGrowth', 1, ...
'displayedGrowth', 1, ...
'displayedMulti', [], ...
'allowNegativeGrowth', true, ...
... % If true, the growth morphogens KAPAR, KAPER, KBPAR, and KBPER
... % can be negative. If false, any negative values arising in the
... % computation are replaced by zero.
'usePrevDispAsEstimate', true, ...
... % If true, the computation of elastic deformation will use the
... % displacements computed on the previous timestep as the initial
... % estimate for the current timestep. If false, the initial estimate
... % will be zero.
'perturbInitGrowthEstimate', 0.00001, ...
... % This defines the amount of random perturbation added to the initial
... % estimate for the computation of elastic deformation, as an absolute amount.
'perturbRelGrowthEstimate', 0.01, ...
... % This defines the amount of random perturbation added to the initial
... % estimate for the computation of elastic deformation, as a relative amount.
'perturbDiffusionEstimate', 0.0001, ...
... % This defines the amount of random perturbation added to the initial
... % estimate for the computation of diffusion.
'resetRand', false, ...
... % If true, the random number generator will be reset to a fixed initial
... % state before each use made of it. This allows exact reproducibility
... % of a simulation. If false, it is never reset.
'mingradient', 0, ...
'relativepolgrad', false, ...
'usefrozengradient', true, ...
'userpolarisation', false, ...
'thresholdsq', 0, ...
'splitmargin', 1.4, ...
... % When splitting of long edges is enabled, an edge will be bisected
... % if its length is this amount times the threshold. The default
... % value is approximately sqrt(2), so that after splitting the two new
... % edges are below the threshold by the same ratio.
'splitmorphogen', , ...
... % The value of this is the name of a morphogen to be used to influence
... % edge-splitting. Every edge where this morphogen exceeds a specified
... % theshold at both ends will be split. See also 'thresholdmgen'.
'thresholdmgen', 0.5, ...
... % This is the threshold value for morphogen-specified edge-splitting.
... % See also 'splitmorphogen'.
'bulkmodulus', 1, ...
... % IGNORE
'unitbulkmodulus', true, ...
... % IGNORE
'poissonsRatio', 0.3, ...
... % This sets Poisson's ratio for the material, a constant value everywhere.
'starttime', 0, ...
... % This defines the time at the start of the simulation.
'timestep', 0.01, ...
... % This is the time step of the simulation. It should be chosen small enough
... % that in a single time step, no part of the mesh grows by more than 10% in
... % any direction, and rotates by no more than 10 degrees. For more accurate
... % runs, these criteriashould be reduced to 5% and 5 degrees.
'timeunitname', , ...
... % This is the name of the unit of time, and should be given in the singular.
'distunitname', 'mm', ...
... % This is the name of the unit of length.
'scalebarvalue', 0, ...
'validateMesh', true, ...
... % This is for debugging only. If true, a large set of validation checks
... % is run after each simulation step, to verify the integrity of the data
... % structures.
'rectifyverticals', false, ...
... % If true, after each simulation step the pairs of vertexes on opposite
... % side of the mesh are moved as necessary to ensure that the line joinging
... % them is perpendicular to the mesh. The midpoint of this line does not move.
... % This should be thought of as revising the decomposition of the material
... % into finite elements, not as a deformation of the material.
... % WARNING
... % BEFORE 2013 this was always treated as true (even if set false).
... % From 2013 onwards it is set true by default.
... %
'allowSplitLongFEM', true, ...
... % This boolean flag specifies whether edges exceeeding a certain length
... % threshold should be split after each simulation step.
'longSplitThresholdPower', 0, ...
'allowSplitBentFEM', false, ...
'allowSplitBio', true, ...
'allowFlipEdges', false, ...
'allowElideEdges', true, ...
'mincellangle', 0.2, ...
'alwaysFlat', false, ...
'flattenforceconvex', true, ...
'flatten', false, ... % IGNORE. Obsolete but harmless
'flattenratio', 1, ... % IGNORE. Obsolete but harmless
'useGrowthTensors', false, ...
'plasticGrowth', false, ...
... % If true, the material is modelled as a fluid flowing with Reynolds
... % number equal to zero, which is the limiting behaviour as Poisson's
... % ratio tends to 0.5. The default is to model it as an elastic solid.
'totalinternalrotation', 0, ... % IGNORE
'stepinternalrotation', 2, ... % IGNORE radians
'showinternalrotation', false, ... % IGNORE
'performinternalrotation', false, ... % IGNORE
'internallyrotated', false, ... % IGNORE
'maxFEcells', int32(0), ...
... % This specifies the maximum number of finite elements the mesh is
... % allowed to contain. When it reaches this number, no more edge-splitting
... % will be done. A value of zero means the number is unlimited.
'inittotalcells', int32(0), ...
'bioApresplitproc', , ...
... % This should be the name of a function which will be called immediately
... % before determining whether any biological cells should be split. This
... % function can be used to override the default cell-splitting method. The
... % method of writing this function is rather complex and not documented here.
'bioApostsplitproc', , ...
... % This should be the name of a function which will be called immediately
... % after having split some biological cells. This
... % function can be used to override the default cell-splitting method. The
... % method of writing this function is rather complex and not documented here.
'maxBioAcells', int32(0), ...
... % This specifies the maximum number of biological cells the mesh is
... % allowed to contain. When it reaches this number, no more cell-splitting
... % will be done. A value of zero means the number is unlimited.
'colors', secondlayercolors, ...
'colorvariation', secondlayercolorvariation, ...
'colorparams', makesecondlayercolorparams( secondlayercolors, secondlayercolorvariation ), ...
'biocolormode', 'auto', ...
'freezing', 0, ...
'canceldrift', false, ...
... % If true, after each simulation step, the mesh is repositioned if necessary
... % to force its centre (defined as the average of all of its vertexes) to
... % remain stationary.
'mgen_interaction', , ...
... % READ-ONLY. This is a handle to the interaction function, and should not be
... % modified by the user.
'mgen_interactionName', , ...
... % READ-ONLY. This is the name of the interaction function, and should not be
... % modified by the user.
'allowInteraction', true, ...
... % This specifies whether the interaction function should be called during each
... % simulation step.
'interactionValid', true, ... % UNSAVED
... % READ-ONLY. If an error is detected in the interaction function, this
... % flag will be set to false. It reverts to true of GFtbox detects that
... % the user has edited it, or when the Reset button in the GUI is clicked.
'gaussInfo', gGAUSS_INFO, ... % IGNORE
'stitchDFs', [], ...
'D', zeros(6,6), ... % IGNORE
'C', zeros(6,6), ... % IGNORE
'G', zeros(6,1), ... % IGNORE
'solver', 'cgs', ...
... % This specifies which solver to user when computing elastic deformation.
'solverprecision', 'double', ...
'solvertolerance', 0.001, ...
'solvertolerancemethod', 'max', ...
'diffusiontolerance', 0.00001, ...
'allowsparse', true, ...
'maxIters', int32(0), ...
'maxsolvetime', 1000, ...
'cgiters', int32(0), ...
'simsteps', int32(0), ...
'stepsperrender', int32(0), ...
'growthEnabled', true, ...
... % This specifies whether elastic growth is to be computed during a
... % simulation step.
'diffusionEnabled', true, ...
... % This specifies whether diffusion is to be computed during a
... % simulation step.
'flashmovie', false, ...
... % This specifies whether all movies should be automatically
... % converted to Flash format on completion.
'makemovie', false, ...
... % This specifies whether a frame is to be saved to a movie file
... % after each simulation step.
'moviefile', , ...
... % The name of the current movie file (empty when a movie is not being recorded).
'codec', 'None', ...
... % The codec to be uesd when recording a movie file.
'autonamemovie', true, ...
... % If true, when a movie is started, a name will be automatically
... % generated for the movie file. If false, the user will be presented
... % with a file dialog to choose the name.
'overwritemovie', false, ...
... % When this is true, no check will be made to see if an automatically
... % generated movie file name already exists. If false, the name will be
... % modified by appending a unique number in order to prevent any existing
... % fie from being overwritten.
'framesize', [], ... % IGNORE UNSAVED
'mov', [], ... % IGNORE UNSAVED
'boingNeeded', false, ... % IGNORE
'initialArea', 0, ...
'bendunitlength', 0, ...
'targetRelArea', 1, ... % UNSAVED
'defaultinterp', 'min', ...
'readonly', false, ...
'projectdir', , ...
'modelname', , ...
'allowsave', true, ... % IGNORE UNSAVED
'addedToPath', false, ... % IGNORE UNSAVED
'bendsplit', 0.3, ...
'usepolfreezebc', false, ...
'dorsaltop', true, ...
'defaultazimuth', -45, ...
'defaultelevation', 33.75, ...
'defaultroll', 0, ...
'defaultViewParams', gMatlabViewParams, ...
'comment', , ...
'legendTemplate', '%T: %q\n%m', ...
... % This is the format string used to define the legend appearing
... % at the top of the picture area.
'bioAsplitcells', true, ...
... % Allow splitting of biological cells.
'bioApullin', 4/28, ...
'bioAfakepull', 0.7/(2*sqrt(3)), ...
'interactive', false, ...
... % READ-ONLY. Says whether GFtbox is running interactively. Obsolete?
'coderevision', int32(0), ...
... % READ-ONLY. The current revision number of GFtbox.
'coderevisiondate', , ...
... % READ-ONLY. The current revision date of GFtbox.
'modelrevision', int32(0), ...
... % READ-ONLY. The latest revision of GFtbox that this model was
... % operated on with. Backward compatibility is retained: old models
... % should always be loadable into new versions of GFtbox. The reverse
... % is not necessarily the case. This field will tell you if the model
... % has been edited with a newer version of GFtbox that you are currently
... % running.
'modelrevisiondate', , ...
... % READ-ONLY. The date of the latest revision of GFtbox that this model was
... % operated on with. See also 'modelrevision'.
'savedrunname', , ... % IGNORE
'savedrundesc', ... % IGNORE
);
gGlobalProps.vxgrad(:,:,1) = gradN( [ 0; 0; 1 ] );
gGlobalProps.vxgrad(:,:,2) = gradN( [ 1; 0; 1 ] );
gGlobalProps.vxgrad(:,:,3) = gradN( [ 0; 1; 1 ] );
gGlobalProps.vxgrad(:,:,4) = gradN( [ 0; 0; -1 ] );
gGlobalProps.vxgrad(:,:,5) = gradN( [ 1; 0; -1 ] );
gGlobalProps.vxgrad(:,:,6) = gradN( [ 0; 1; -1 ] );
gGlobalDynamicProps = struct( ... % ITEM leaf_setproperties
'currenttime', 0.0, ... % DYNAMIC
'currentIter', 0, ... % DYNAMIC
'laststagesuffix', , ... % DYNAMIC, UNSAVED
'currentArea', 0, ... % DYNAMIC
'previousArea', 0, ... % DYNAMIC
'thicknessAbsolute', 0, ... % DYNAMIC?
'cellscale', 0, ... % DYNAMIC
'locatenode', 0, ... % DYNAMIC
'locateDFs', [], ... % DYNAMIC
'staticreadonly', false, ... % DYNAMIC
'commandok', true, ... % DYNAMIC
'doinit', true ... % DYNAMIC
);
gOurViewParams = struct( ...
'azimuth', -45, ...
'elevation', 33.75, ...
'roll', 0, ...
'fov', 10, ...
'pan', [0 0], ...
'targetdistance', 0, ...
'camdistance', 4*sqrt(3)/(2*tan(5*pi/180)), ...
'projection', 'orthographic' );
gMatlabViewParams = cameraParamsFromOurViewParams( gOurViewParams );
gOurViewParamNames = fieldnames( gOurViewParams );
gMatlabViewParamNames = fieldnames( gMatlabViewParams );
gDefaultPlotOptions = struct( ... % ITEM leaf_plotoptions
'drawtensoraxes', false, ...
'unitcrosses', true, ...
... % 'tensorquantity', 'resultantgrowthrate', ... % New 2009-11-11
... % 'tensorproperty', 'total', ...
'outputquantity', 'resultantgrowthrate', ...
'outputaxes', 'total', ...
'morphogen', 1, ...
... % 'rawstuff', [], ... % Obsolete.
'drawleaf', 1, ...
'drawedges', 1, ...
'drawseams', 1, ...
'fillleaf', 1, ...
'drawgradients', 0, ...
'unitgradients', 0, ...
'multibrighten', 0.1, ...
... % 'monochrome', true, ...
'drawcolorbar', true, ...
'monocolors', [[0 0 1];[1 0 0]], ...
'canvascolor', [1 1 1], ...
'cmap', [], ...
'crange', [], ...
'zerowhite', 0, ...
'cmaptype', 'monochrome', ...
'sparsedistance', 0, ...
'staticdecor', true, ...
'axisRange', [], ...
'axisVisible', 1, ...
'alpha', 0.8, ...
'ambientstrength', 0.8, ...
'azimuth', -45, ...
'elevation', 33.75, ...
'roll', 0, ...
'ourViewParams', gOurViewParams, ...
'matlabViewParams', gMatlabViewParams, ...
'autozoom', false, ...
'autocentre', false, ...
'autoScale', 1, ...
'autoColorRange', 1, ...
'drawsecondlayer', 0, ...
'cellsonbothsides', true, ...
'layeroffset', 0.2, ...
'FEthicklinesize', 2, ...
'FEthinlinesize', 1, ...
'FElinecolor', [0 0 0], ...
'seamlinesize', 5, ...
'seamlinecolor', [1 0 0], ...
'bioAlinesize', 1, ...
'bioAlinecolor', [0.2 0.2 0.2], ...
'bioAnewlinecolor', [0.2 0.2 0.2], ...
'bioApointsize', 3, ...
'bioApointcolor', [0.2 0.2 0.2], ...
'bioAnewpointcolor', [0.2 0.2 0.2], ...
'bioAalpha', 1, ...
'axescolor', [0 0 0.3], ...
'decorateAside', false, ...
'drawnormals', 0, ...
'drawmutant', 1, ...
'drawdisplacements', 0, ...
'drawlegend', 1, ...
'drawscalebar', 1, ...
'thick', 1, ...
'nodenumbering', 0, ...
'nodenumbercolor', [1 0 0], ...
'edgenumbering', 0, ...
'edgenumbercolor', [0 0.4 0.1], ...
'FEnumbering', 0, ...
'FEnumbercolor', [0 0 0.4], ...
'texture', 0, ...
'bgcolor', [1 1 1], ...
'emptycolor', [0.7, 0.8, 1], ...
'clippingAzimuth', 0, ...
'clippingElevation', 0, ...
'clippingDistance', -0.01, ...
'clipbymgen', false, ...
'clipmgens', [], ...
'clipmgenthreshold', 0, ...
'clipmgenabove', true, ...
'clipmgenall', true, ...
'doclip', false, ...
'uicontrols', true, ...
'light', false, ...
'lightmode', 'gouraud', ...
'invisibleplot', false, ...
'decorscale', 1, ...
'highlightthickness', 3, ...
'arrowthickness', 2, ...
'crossthickness', 1, ...
'arrowheadsize', 0.5, ...
'arrowheadratio', 0.3, ...
'highgradcolor', [0 0 0.3], ...
'lowgradcolor', [0.5 0 0], ...
'hiresdpi', 400, ...
'mgenownsideonly', 'false', ...
'taper', 'true', ...
'anisotropythreshold', 0, ...
'linesmoothing', 'on', ...
'userplotproc', [] );
end