Research: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
=<span style="color:DarkGreen;">Computational biology</span>= | |||
---- | |||
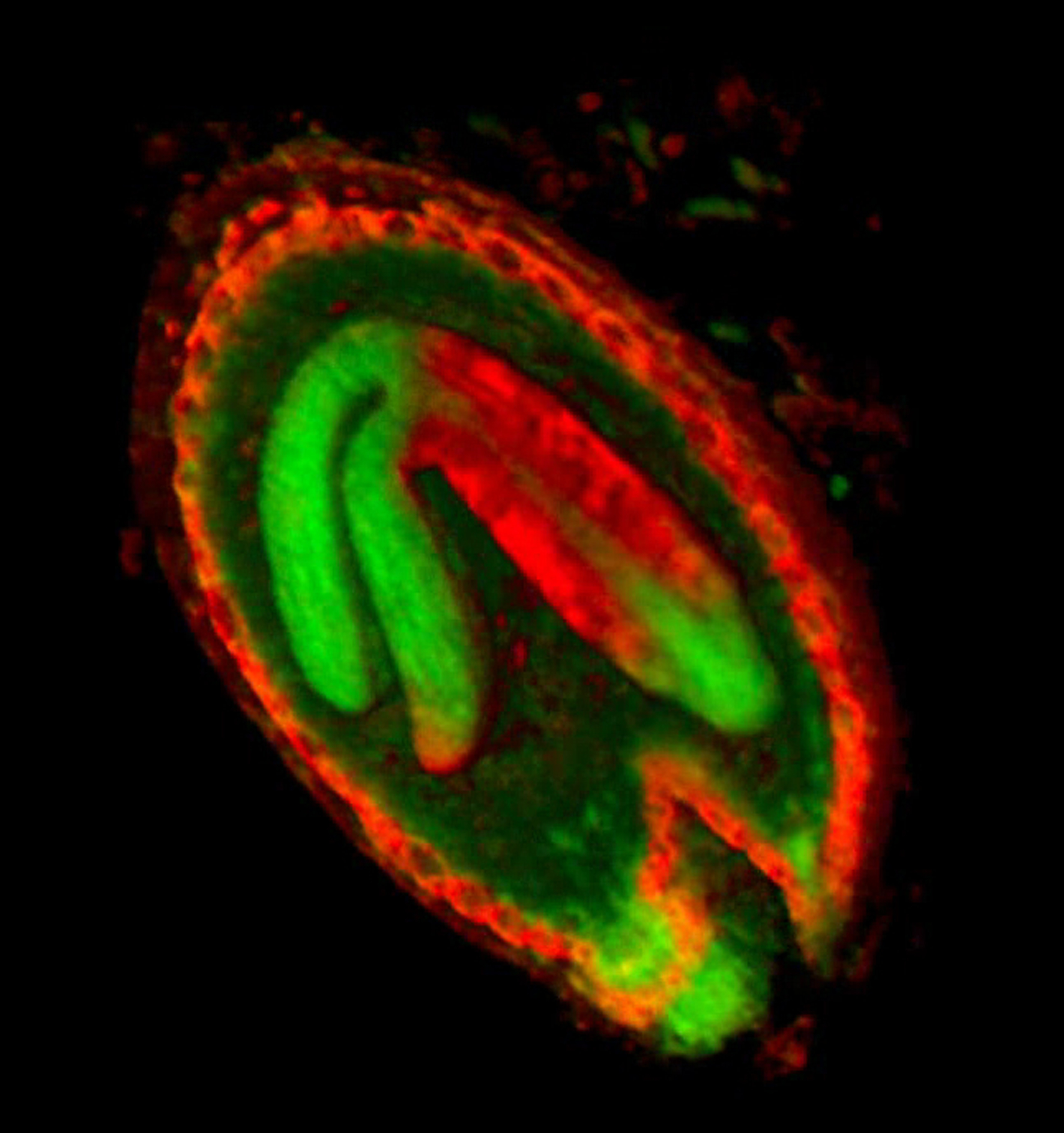
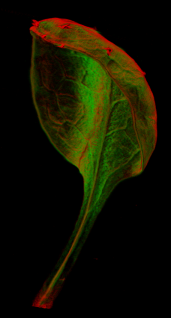
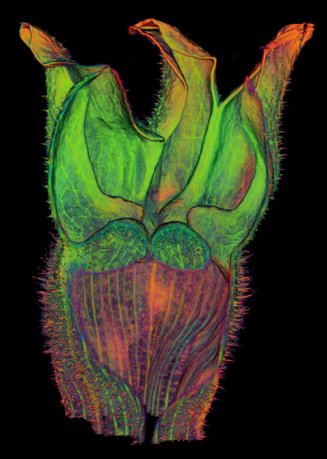
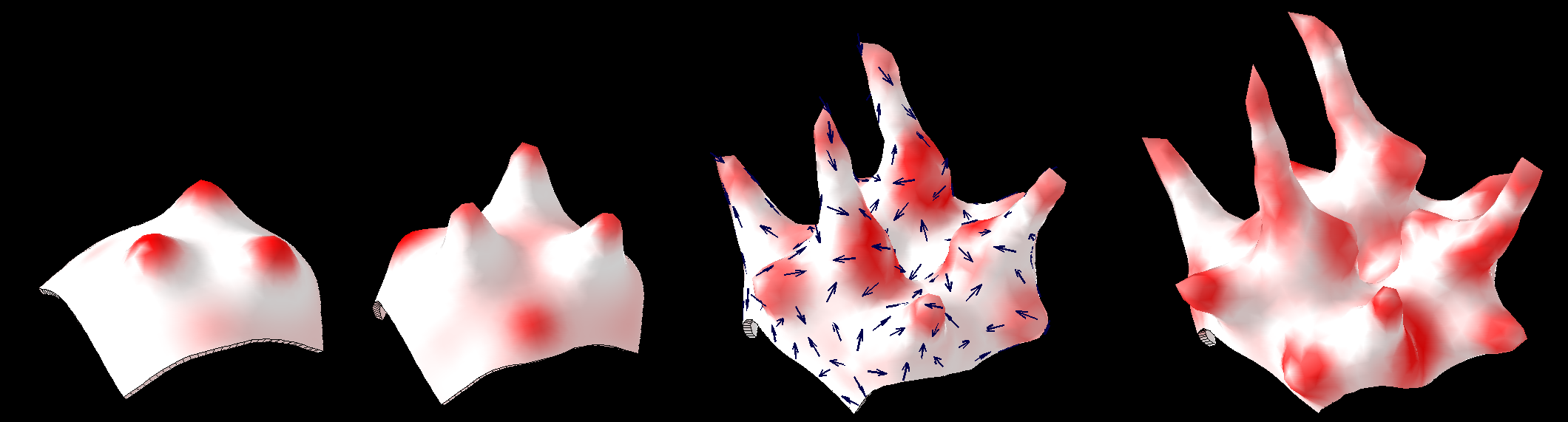
{|border="0" | ==<span style="color:DarkGreen;">[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;"> '''Growing''']] complex biological shapes from patterns of gene expression</span>== | ||
|align="center"| | {| border="0" width=100% style="background-color:#000000;" | ||
|- | |||
| | |align="center"| | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000340-0001.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000490-0001.png|120px]] | |||
< | [[Image:LabelledCropped GPT Snapdragon 2010-000570-0002.png|120px]] | ||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0007.png|120px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0003 double.png|100px]] | |||
[[Image:LabelledCropped GPT Snapdragon 2010-000570-0002 triple.png|120px]] | |||
|} | |||
<br> | |||
[[Software#Quantitative understanding of growing shapes: GFtbox|<span style="color:Green;">'''MORE'''</span>]]<br> | |||
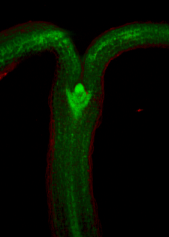
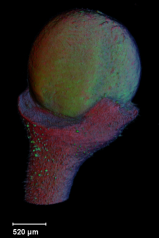
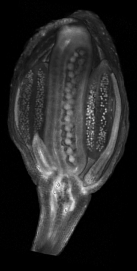
==<span style="color:DarkGreen;">[[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;"> '''Viewing''']] three dimensional volume (microscopy) images== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |align="center"| | ||
[[Image:Cs0prxz0.png|32x32px]] | |||
| | [[Image:Leaf_trichomes.png|50px]] | ||
[[Image:Cs0prxz0.png|50px]] | |||
| | [[Image:GL2_GUS.png|50px]] | ||
[[Image:Leaf5.png|50px]] | |||
[[Image:OleosinSeed.png|50px]] | |||
[[Image:OPT_Leaf_copy.png|50px]] | |||
[[Image:Seedling_copy.png|50px]] | |||
[[Image:Snapdragon_Peloric_mutant.png|50px]] | |||
[[Image:Tissue.png|50px]] | |||
[[Image:Z9r3j2yx.png|50px]] | |||
[[Image:1896_wh_txr_light.png|50px]] | |||
[[Image:Ara_flower.png|50px]] | |||
[[Image:Arableaf_ath8_OPT.png|50px]] | |||
|} | |} | ||
<br> | |||
[[Software#Viewing and measuring volume images: VolViewer|<span style="color:Green;">'''MORE'''</span>]] | |||
=''' | ==[[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''Analysing'''</span>]] shapes: faces, leaves and flowers== | ||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
[[Image:PortraitsMEANSsmaller.jpg|800px]] | |||
|-} | |||
<br> | |||
[[Software#Analysing shapes in 2D and 3D: AAMToolbox|<span style="color:Green;">'''MORE'''</span>]]<br> | |||
Have you seen the original paintings? Do they exist?. <br><br> | |||
=<span style="color:Navy;">Algorithms= | |||
==''' | ---- | ||
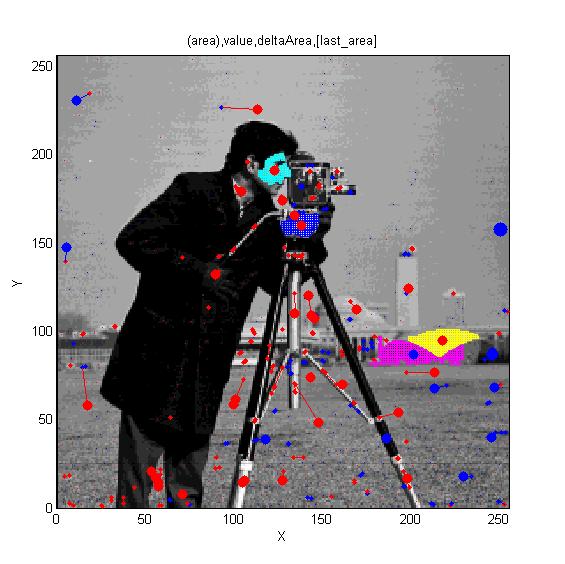
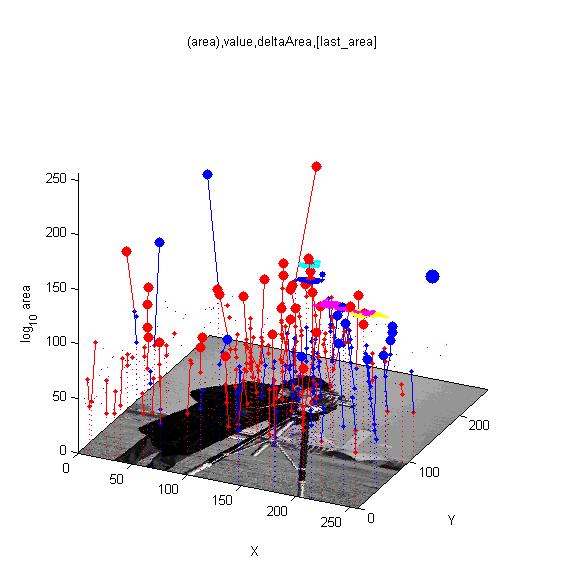
==[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#Art.2C_extrema_of_light_and_shade:_PhotoArtMaster<span style="color:Navy;">'''Vision''':] MSER's, extrema, filter-banks, Sieves and '''Scale-space'''== | |||
{| border="0" width=100% style="background-color:#ffffff;" | |||
|- | |||
|align="center"| | |||
[[Image:Cameraman_iso_topview.jpg|300px|AAMToolbox]] | |||
[[Image:Cameraman_iso_tree.jpg|300px|AAMToolbox]] | |||
|} | |||
[[ | [[Software#MSERs.2C_extrema.2C_connected-set_filters_and_sieves|<span style="color:Navy;">'''MORE'''</span>]] | ||
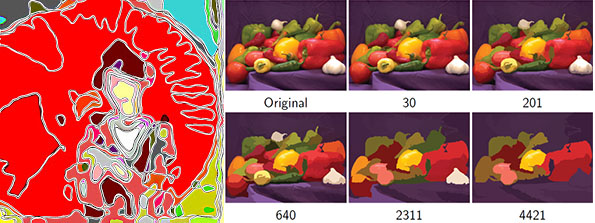
''' | ==[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#Art.2C_extrema_of_light_and_shade:_PhotoArtMaster <span style="color:Navy;">'''Applications'''</span>]' <span style="color:Navy;">of non-linear filter banks (sieves) and the art of light and shade</span>== | ||
{| border="0" width=100% style="background-color:#ffffff;" | |||
|- | |||
|align="center"| | |||
[[Image:Colour_sieve.jpg|600px|AAMToolbox]] | |||
|} | |||
These images were produced from photographs using '''ArtMaster''' (formally known as '''PhotoArtMaster'''). The software received many favourable reviews when it was released (e.g. [http://graphicssoft.about.com/cs/photoart/gr/photoartmasterg.htm "This software can give you a lot of satisfaction from your everyday photos"], [http://graphicssoft.about.com/library/products/aafpr_photoartmaster1.htm] | |||
The | [http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#The_final_version_of_the_Windows_ArtMaster2.0_is_downloadable_here_with_no_support The final (so far unpublished) version of ArtMaster including code is downloadable from here.] I cannot provide support but quite of lot of documentation is available within [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this document''''' </span>] | ||
[http://cmpdartsvr3.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software#Art.2C_extrema_of_light_and_shade:_PhotoArtMaster <span style="color:Navy;">'''MORE'''</span>] | |||
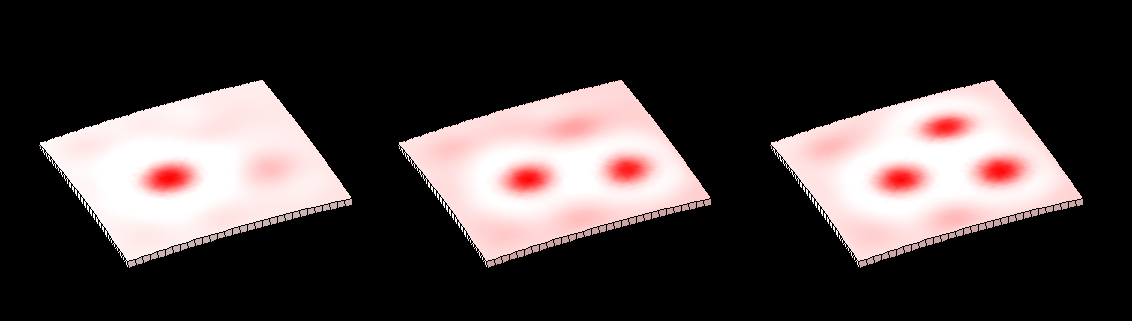
==[[Software#Reaction-diffusion and morphogenesis| <span style="color:Navy;"> '''Reaction-diffusion'''</span>]] <span style="color:Navy;">and morphogenesis - the growth of shapes== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
[[Image:tentacles_reaction_diffusion.png|400px]] | |||
[[Image:tentacles_morphogenesis.png|600px]] | |||
< | |} | ||
This image forms part of a 'journey' in the Science Museum of London's 'Journeys of Invention' [http://www.sciencemuseum.org.uk/journeys iPad app.]<br><br> | |||
[[Software#Reaction-diffusion and morphogenesis|<span style="color:Navy;">'''MORE'''</span>]]<br><br> | |||
= | |||
[[Image: | |||
< | |||
<br | |||
Latest revision as of 11:57, 14 April 2014
Computational biology
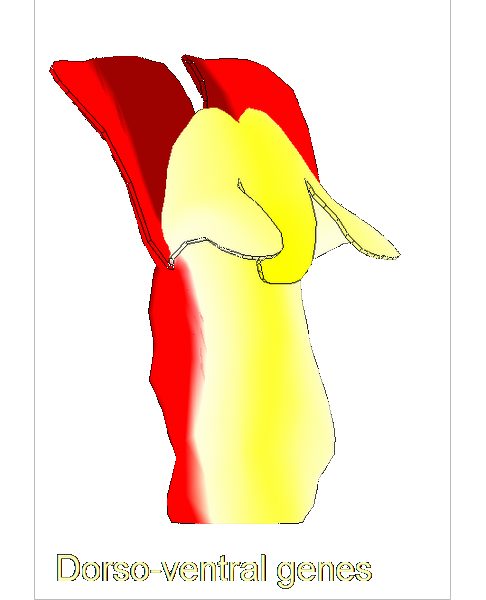
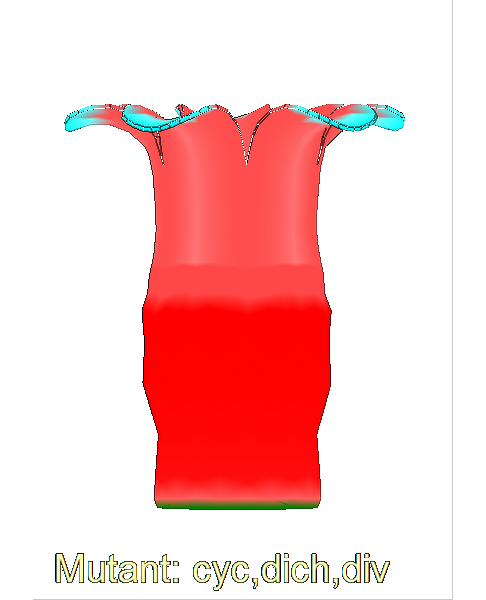
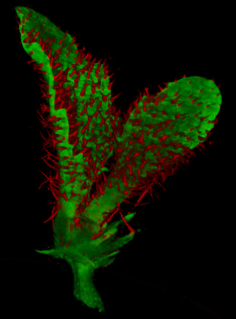
Growing complex biological shapes from patterns of gene expression
Viewing three dimensional volume (microscopy) images
Analysing shapes: faces, leaves and flowers

MORE
Have you seen the original paintings? Do they exist?.
Algorithms
Vision: MSER's, extrema, filter-banks, Sieves and Scale-space
Applications' of non-linear filter banks (sieves) and the art of light and shade
These images were produced from photographs using ArtMaster (formally known as PhotoArtMaster). The software received many favourable reviews when it was released (e.g. "This software can give you a lot of satisfaction from your everyday photos", [1]
The final (so far unpublished) version of ArtMaster including code is downloadable from here. I cannot provide support but quite of lot of documentation is available within 'this document
Reaction-diffusion and morphogenesis - the growth of shapes
This image forms part of a 'journey' in the Science Museum of London's 'Journeys of Invention' iPad app.
MORE