SectorAnalysisToolbox Documentation: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
'''How to use the tutorial.''' First [[SectorAnalysisToolbox Download|download and install the SectorAnalysisToolbox ]]. Then, from Matlab, change directory into the project and launch the SectorAnalysisToolbox. | '''How to use the tutorial.''' First [[SectorAnalysisToolbox Download|download and install the SectorAnalysisToolbox ]]. Then, from Matlab, change directory into the project and launch the SectorAnalysisToolbox. | ||
<gallery caption="antirrhinum petals" widths="200px" heights="100px" perrow="8" border="0"> | <gallery caption="antirrhinum petals sample data" widths="200px" heights="100px" perrow="8" border="0"> | ||
Image:P9F17P1I.JPG | Image:P9F17P1I.JPG | ||
Image:P9F17P2I.JPG | Image:P9F17P2I.JPG | ||
Revision as of 16:03, 9 March 2012
The models shown in these tutorials illustrate features of the SectorAnalysisToolbox software. Viewing these pages. Some versions of Firefox and Explorer do not create satisfactory prints even though you can view the pages with no problems. Chrome does appear to produce good printouts.
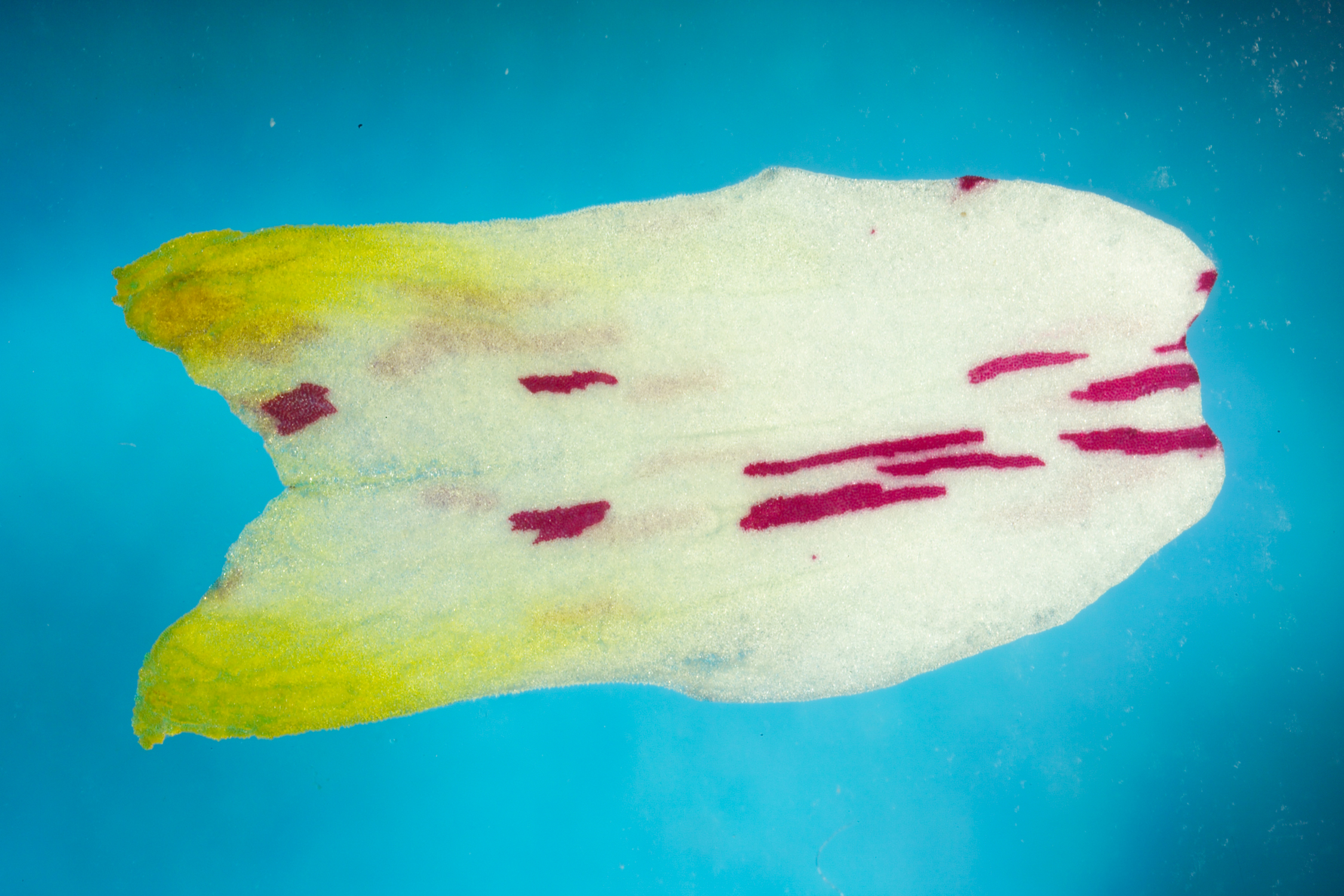
Using the Sector Analysis Toolbox (SAT) to analyse the shapes of cell clones in antirrhinum petals
How to use the tutorial. First download and install the SectorAnalysisToolbox . Then, from Matlab, change directory into the project and launch the SectorAnalysisToolbox.
- antirrhinum petals sample data
1 Creating a new project
To create a new project, click the New Project button and name your project when prompted. The toolbox automatically gives each project the prefix ‘SAP_’ (Sector Analysis Project). The project directory structure will be saved to disk and contains the folders Original, Processed and Staged. Copy the sample antirrhinum images (P9F17P1I.JPG, P9F17P2I.JPG, P10F17P1I.JPG) into the Staged dir.
Once a project has been created, you can return to that project either by selecting that project in the ‘Current Directory’ line at the top of the Matlab window, or by opening the toolbox and using the Browse button to select your project.
2 Segmenting the petal and clones
We have created a new project and populated it with some sample antirrhinum petal images. We would now like to automatically segment out the antirrhinum petal and the purple sectors.
Click the button Segment Images. A dialog box will ask the user to select a segmentation algorithm. Select amelia_petal_sectors.m from the drop down box as this segmentation algorithm is specifically designed to work with our data. The SAT will look in the Staged directory for the *.jpg imaged to segment. A dialog box will ask the user which images to segment - in this case we will use the Select All button. A task bar indicates segmentation progress. When an image has been segmented, the image file in the ‘Staged’ folder is replaced by a folder with containing a matlab file called ‘edge_xy’ (the organ outline coordinates), a .jpg snapshot of the original image, a .tif black and white (binary) image with the suffix ‘_organ’ (corresponding to the petal/leaf outline), and a .tif binary image with the suffix ‘_sectors’ (corresponding to the sectors).
- The original image (left) segmented petal (centre) and segmented clones (right)
3 Editing the Segmented clones and organ edges
Click Sector Editor to open the Sector Editor Tool (see image to the right). The first image from the ‘Staged’ folder should be displayed in this window. The outline of the petal section is marked by 50 edge points joined by a red line. Edge information are provided by the edge_xy.mat file and filename_organ.tif. The latter shows the organ shape as a binary (black and white) image. If this shape is not accurate due to, e.g. an air bubble, the egde points will not follow the organ shape exactly. If you find that most points are too far off consider using a differnt segmentation algorithm. The file filename_sectors.tif is openend (the segemented out sectors) to superimpose the sectors on the image in blue. The boxes Show Sectors and Show Edge can be ticked or made blank to show or hide the edge outline and the sector pattern found by the computer. Again, if sectors are not recignised properly, change the segementation algorithm. The tools at the top of the window can be used to zoom into, pan around and rotate the image (standard symbols are used for each function). Edges can be moved using the ‘Move Edge Points’ button. To smooth the edges, click on “Def. primary points” and select at least 2 primaries (for example tip and base: these must correspond to the same number and the same morphological feature for each image). Then click “smooth pts”, all the edge points will be smoothed around the 2 fixed primaries (these will not move). It is important that the edges are consistent from one image to another as they will be used to calculate the mean shape and the warping of the sectors. When the edge points are judged to capture the petal outline satisfactorily, the same button (now named ‘Save New Edge’) should be clicked to save the edited outline under edge_xy.may. The primary points are saved under primary_landmarks.mat.