Tutorial on two romatic hearts
Back to GFtbox Tutorial pages
We illustrate the practical advantage of having submodels within a project and an important consequence of understanding biological growth within the GPT-framework.
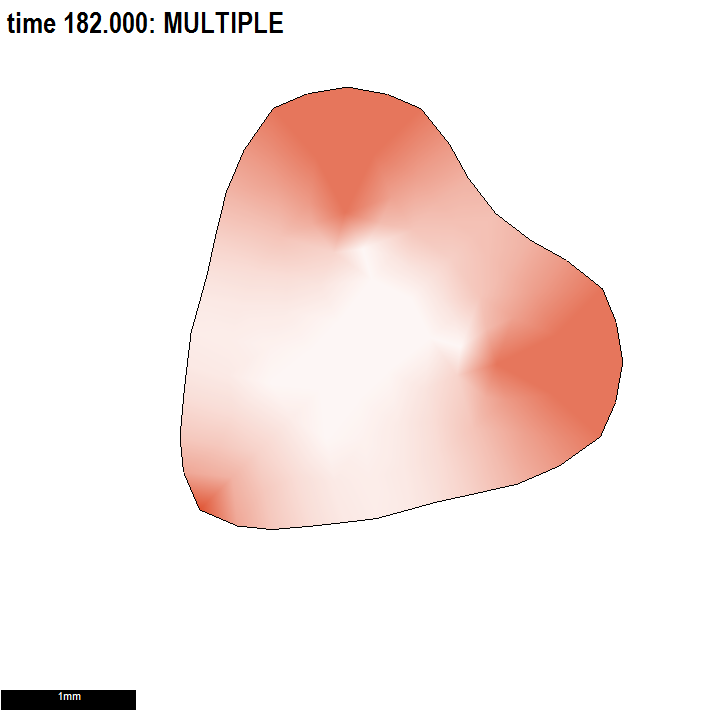
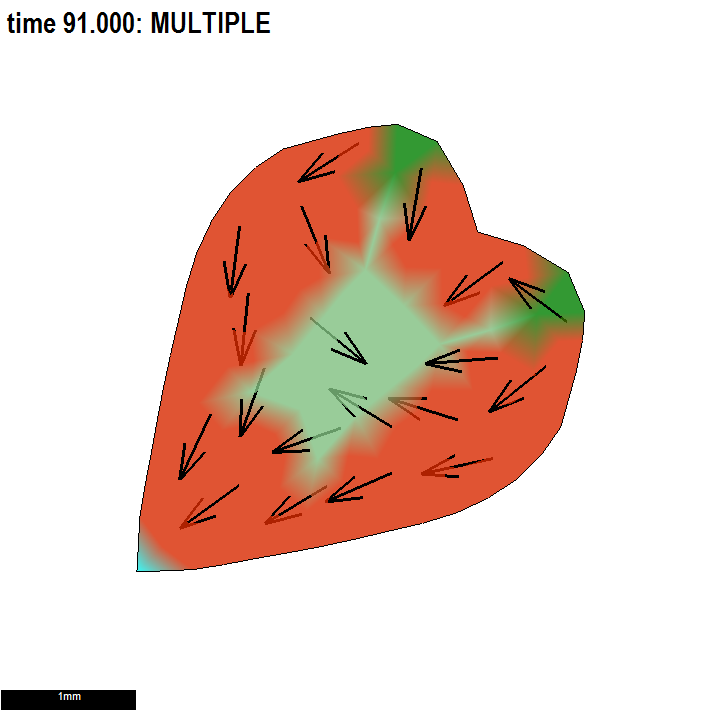
The aim is to grow a disc into a romantic heart
Interaction function in detail (A newly created, empty, interaction function is shown here.)
Section 1 function m = gpt_twowayheart_20110531( m ) %m = gpt_twowayheart_20110531( m ) % Morphogen interaction function. % Written at 2011-05-31 19:51:32. % GFtbox revision 3548, 2011-05-31 14:37:10.747930. % The user may edit any part of this function between delimiters % of the form "USER CODE..." and "END OF USER CODE...". The % delimiters themselves must not be moved, edited, deleted, or added. if isempty(m), return; end fprintf( 1, '%s found in %s\n', mfilename(), which(mfilename()) ); try m = local_setproperties( m ); catch end realtime = m.globalDynamicProps.currenttime; Section 2 %%% USER CODE: INITIALISATION % In this section you may modify the mesh in any way whatsoever. if (Steps(m)==0) && m.globalDynamicProps.doinit % First iteration % Set up names for variant models. Useful for running multiple models on a cluster. m.userdata.ranges.modelname.range = { 'PolariserBased', 'DifferentialGrowthBased' }; % CLUSTER m.userdata.ranges.modelname.index = 1; % CLUSTER end modelname = m.userdata.ranges.modelname.range{m.userdata.ranges.modelname.index}; % CLUSTER disp(sprintf('\nRunning %s model %s\n',mfilename, modelname)); % Set priorities for simultaneous plotting of multiple morphogens, if desired. m = leaf_mgen_plotpriority( m, {'ID_PLUSORG', 'ID_MINUSORG'}, [1,2], [0.4,0.4] ); % Set colour of polariser gradient arrows. m = leaf_plotoptions(m,'highgradcolor',[0,0,0],'lowgradcolor',[1,0,0]); m = leaf_plotoptions(m,'decorscale',1.5); % setup a multiplot of the following morphogens m = leaf_plotoptions( m, 'morphogen', {'V_KAREAL','ID_PLUSORG','ID_MINUSORG'}); %%% END OF USER CODE: INITIALISATION Section 3 %%% SECTION 1: ACCESSING MORPHOGENS AND TIME. %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. if isempty(m), return; end setGlobals(); global gNEW_KA_PAR gNEW_KA_PER gNEW_KB_PAR gNEW_KB_PER global gNEW_K_NOR gNEW_POLARISER gNEW_STRAINRET gNEW_ARREST dt = m.globalProps.timestep; polariser_i = gNEW_POLARISER; P = m.morphogens(:,polariser_i); [kapar_i,kapar_p,kapar_a,kapar_l] = getMgenLevels( m, 'KAPAR' ); [kaper_i,kaper_p,kaper_a,kaper_l] = getMgenLevels( m, 'KAPER' ); [kbpar_i,kbpar_p,kbpar_a,kbpar_l] = getMgenLevels( m, 'KBPAR' ); [kbper_i,kbper_p,kbper_a,kbper_l] = getMgenLevels( m, 'KBPER' ); [knor_i,knor_p,knor_a,knor_l] = getMgenLevels( m, 'KNOR' ); [strainret_i,strainret_p,strainret_a,strainret_l] = getMgenLevels( m, 'STRAINRET' ); [arrest_i,arrest_p,arrest_a,arrest_l] = getMgenLevels( m, 'ARREST' ); [id_plusorg_i,id_plusorg_p,id_plusorg_a,id_plusorg_l] = getMgenLevels( m, 'ID_PLUSORG' ); [id_minusorg_i,id_minusorg_p,id_minusorg_a,id_minusorg_l] = getMgenLevels( m, 'ID_MINUSORG' ); [v_kareal_i,v_kareal_p,v_kareal_a,v_kareal_l] = getMgenLevels( m, 'V_KAREAL' ); [id_tip_i,id_tip_p,id_tip_a,id_tip_l] = getMgenLevels( m, 'ID_TIP' ); [id_top_i,id_top_p,id_top_a,id_top_l] = getMgenLevels( m, 'ID_TOP' ); [s_growth_i,s_growth_p,s_growth_a,s_growth_l] = getMgenLevels( m, 'S_GROWTH' ); [id_mid_i,id_mid_p,id_mid_a,id_mid_l] = getMgenLevels( m, 'ID_MID' ); % Mesh type: circle % centre: 0 % circumpts: 24 % coneangle: 0 % dealign: 0 % height: 0 % innerpts: 0 % randomness: 0.1 % rings: 4 % version: 1 % xwidth: 2 % ywidth: 2 % Morphogen Diffusion Decay Dilution Mutant % ------------------------------------------------- % KAPAR ---- ---- ---- ---- % KAPER ---- ---- ---- ---- % KBPAR ---- ---- ---- ---- % KBPER ---- ---- ---- ---- % KNOR ---- ---- ---- ---- % POLARISER 0.1 ---- ---- ---- % STRAINRET ---- ---- ---- ---- % ARREST ---- ---- ---- ---- % ID_PLUSORG ---- ---- ---- ---- % ID_MINUSORG ---- ---- ---- ---- % V_KAREAL ---- ---- ---- ---- % ID_TIP ---- ---- ---- ---- % ID_TOP ---- ---- ---- ---- % S_GROWTH 0.01 ---- ---- ---- % ID_MID ---- ---- ---- ---- %%% USER CODE: MORPHOGEN INTERACTIONS % In this section you may modify the mesh in any way that does not Section 4 % alter the set of nodes. % Use the same pattern for both submodels RangeTip=(m.nodes(:,1)<-0.8)&... (abs(m.nodes(:,2))<0.2); RangeMid=(m.nodes(:,1)<=0.5)&... (m.nodes(:,1)>-0.5)&... (abs(m.nodes(:,2))<0.3); RangeTops=(m.nodes(:,1)<=max(m.nodes(:,1))&... (m.nodes(:,1)>0.5)&... (abs(m.nodes(:,2))>0.3)); if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. switch modelname case 'PolariserBased' % % One way to set up a morphogen gradient is by ... % Setting up a gradient by clamping the ends (execute only once) P(RangeTip)=0; P(RangeMid)=0.5; P(RangeTops)=1; id_plusorg_p=P; id_minusorg_p(RangeTip)=1; m.morphogenclamp( RangeTops|RangeTip|RangeMid, polariser_i ) = 1; m = leaf_mgen_conductivity( m, 'POLARISER', 0.1 ); %specifies the diffusion rate of polariser m = leaf_mgen_absorption( m, 'POLARISER', 0.0 ); % specifies degradation rate of polariser case 'DifferentialGrowthBased' % P(:)=0; % One way to set up a morphogen gradient is by ... % Setting up a gradient by clamping the ends (execute only once) s_growth_p(RangeTip)=1; s_growth_p(RangeMid)=0.05; s_growth_p(RangeTops)=0.8; m.morphogenclamp( RangeTops|RangeTip|RangeMid, s_growth_i ) = 1; m = leaf_mgen_conductivity( m, 's_growth', 0.001 ); %specifies the diffusion rate of polariser m = leaf_mgen_absorption( m, 's_growth', 0.0 ); % specifies degradation rate of polariser end end BasicGrowth=0.01; switch modelname case 'PolariserBased' % % Every equation to be formatted should end with an at-at Eqn N comment. kapar_p(:) = BasicGrowth; % when isotropic this will be 0.005 kaper_p(:) = 0.0; % when isotropic this will be 0.005 kbpar_p(:) = BasicGrowth; % when isotropic this will be 0.005 kbper_p(:) = 0.0; % when isotropic this will be 0.005 knor_p(:) = 0; % thickness case 'DifferentialGrowthBased' % % Every equation to be formatted should end with an at-at Eqn N comment. kapar_p(:) = BasicGrowth*s_growth_p; %0.01; % when isotropic this will be 0.005 kaper_p(:) = BasicGrowth*s_growth_p; %0.0; % when isotropic this will be 0.005 kbpar_p(:) = BasicGrowth*s_growth_p; %0.01; % when isotropic this will be 0.005 kbper_p(:) = BasicGrowth*s_growth_p; %0.0; % when isotropic this will be 0.005 knor_p(:) = 0; % thickness end v_kareal_p=kapar_p+kaper_p; % total specified areal growth Section 5 %%% END OF USER CODE: MORPHOGEN INTERACTIONS %%% SECTION 3: INSTALLING MODIFIED VALUES BACK INTO MESH STRUCTURE %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. m.morphogens(:,polariser_i) = P; m.morphogens(:,kapar_i) = kapar_p; m.morphogens(:,kaper_i) = kaper_p; m.morphogens(:,kbpar_i) = kbpar_p; m.morphogens(:,kbper_i) = kbper_p; m.morphogens(:,knor_i) = knor_p; m.morphogens(:,strainret_i) = strainret_p; m.morphogens(:,arrest_i) = arrest_p; m.morphogens(:,id_plusorg_i) = id_plusorg_p; m.morphogens(:,id_minusorg_i) = id_minusorg_p; m.morphogens(:,v_kareal_i) = v_kareal_p; m.morphogens(:,id_tip_i) = id_tip_p; m.morphogens(:,id_top_i) = id_top_p; m.morphogens(:,s_growth_i) = s_growth_p; m.morphogens(:,id_mid_i) = id_mid_p; %%% USER CODE: FINALISATION %%% END OF USER CODE: FINALISATION end %%% USER CODE: SUBFUNCTIONS Section 6 function m = local_setproperties( m ) end