Tutorial on two romatic hearts
Back to GFtbox Tutorial pages
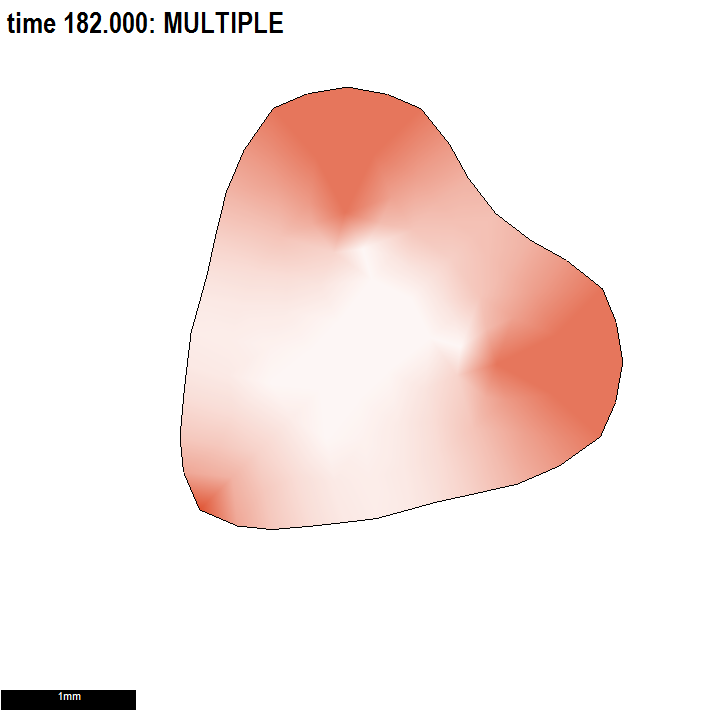
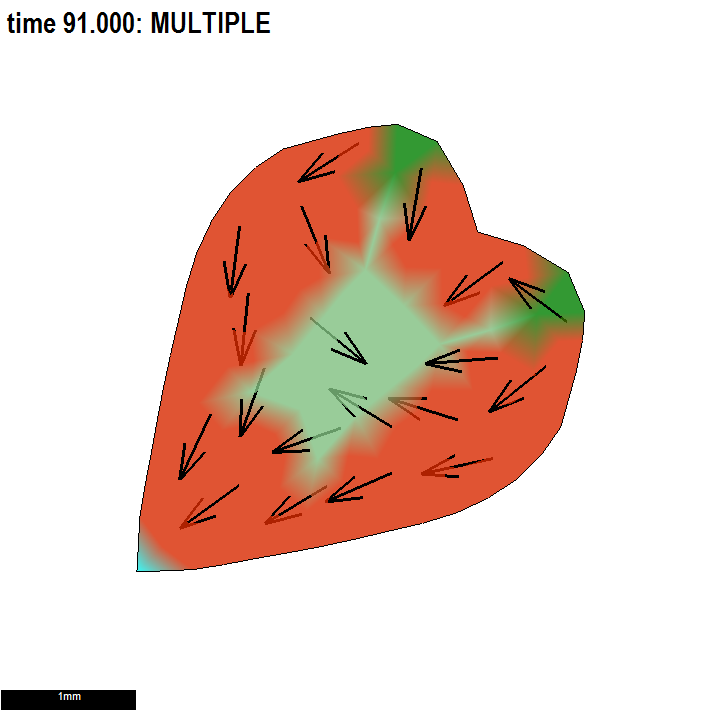
We illustrate the practical advantage of having submodels within a project and an important consequence of understanding biological growth within the GPT-framework.
Conclusion: using a combination of polarity patterns to set local axes for anisotropic growth and patterns of differential specified growth to regulate the growth of shape would be powerful.
Illustrating two independent ways to form shapes and the use of submodels.
The full interaction function is shown at the bottom. The line of code that selects the submodel and the start of each submodel is shown in red.
Section 1 function m = gpt_twowayheart_20110531( m ) %m = gpt_twowayheart_20110531( m ) % Morphogen interaction function. % Written at 2011-05-31 19:51:32. % GFtbox revision 3548, 2011-05-31 14:37:10.747930. % The user may edit any part of this function between delimiters % of the form "USER CODE..." and "END OF USER CODE...". The % delimiters themselves must not be moved, edited, deleted, or added. if isempty(m), return; end fprintf( 1, '%s found in %s\n', mfilename(), which(mfilename()) ); try m = local_setproperties( m ); catch end realtime = m.globalDynamicProps.currenttime; Section 2 %%% USER CODE: INITIALISATION % In this section you may modify the mesh in any way whatsoever. if (Steps(m)==0) && m.globalDynamicProps.doinit % First iteration % Set up names for variant models. Useful for running multiple models on a cluster. m.userdata.ranges.modelname.range = { 'PolariserBased', 'DifferentialGrowthBased' }; % CLUSTER m.userdata.ranges.modelname.index = 1; % CLUSTER end modelname = m.userdata.ranges.modelname.range{m.userdata.ranges.modelname.index}; % CLUSTER disp(sprintf('\nRunning %s model %s\n',mfilename, modelname)); % Set priorities for simultaneous plotting of multiple morphogens, if desired. m = leaf_mgen_plotpriority( m, {'ID_PLUSORG', 'ID_MINUSORG'}, [1,2], [0.4,0.4] ); % Set colour of polariser gradient arrows. m = leaf_plotoptions(m,'highgradcolor',[0,0,0],'lowgradcolor',[1,0,0]); m = leaf_plotoptions(m,'decorscale',1.5); % setup a multiplot of the following morphogens m = leaf_plotoptions( m, 'morphogen', {'V_KAREAL','ID_PLUSORG','ID_MINUSORG'}); %%% END OF USER CODE: INITIALISATION Section 3 %%% SECTION 1: ACCESSING MORPHOGENS AND TIME. %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. if isempty(m), return; end setGlobals(); global gNEW_KA_PAR gNEW_KA_PER gNEW_KB_PAR gNEW_KB_PER global gNEW_K_NOR gNEW_POLARISER gNEW_STRAINRET gNEW_ARREST dt = m.globalProps.timestep; polariser_i = gNEW_POLARISER; P = m.morphogens(:,polariser_i); [kapar_i,kapar_p,kapar_a,kapar_l] = getMgenLevels( m, 'KAPAR' ); [kaper_i,kaper_p,kaper_a,kaper_l] = getMgenLevels( m, 'KAPER' ); [kbpar_i,kbpar_p,kbpar_a,kbpar_l] = getMgenLevels( m, 'KBPAR' ); [kbper_i,kbper_p,kbper_a,kbper_l] = getMgenLevels( m, 'KBPER' ); [knor_i,knor_p,knor_a,knor_l] = getMgenLevels( m, 'KNOR' ); [strainret_i,strainret_p,strainret_a,strainret_l] = getMgenLevels( m, 'STRAINRET' ); [arrest_i,arrest_p,arrest_a,arrest_l] = getMgenLevels( m, 'ARREST' ); [id_plusorg_i,id_plusorg_p,id_plusorg_a,id_plusorg_l] = getMgenLevels( m, 'ID_PLUSORG' ); [id_minusorg_i,id_minusorg_p,id_minusorg_a,id_minusorg_l] = getMgenLevels( m, 'ID_MINUSORG' ); [v_kareal_i,v_kareal_p,v_kareal_a,v_kareal_l] = getMgenLevels( m, 'V_KAREAL' ); [id_tip_i,id_tip_p,id_tip_a,id_tip_l] = getMgenLevels( m, 'ID_TIP' ); [id_top_i,id_top_p,id_top_a,id_top_l] = getMgenLevels( m, 'ID_TOP' ); [s_growth_i,s_growth_p,s_growth_a,s_growth_l] = getMgenLevels( m, 'S_GROWTH' ); [id_mid_i,id_mid_p,id_mid_a,id_mid_l] = getMgenLevels( m, 'ID_MID' ); % Mesh type: circle % centre: 0 % circumpts: 24 % coneangle: 0 % dealign: 0 % height: 0 % innerpts: 0 % randomness: 0.1 % rings: 4 % version: 1 % xwidth: 2 % ywidth: 2 % Morphogen Diffusion Decay Dilution Mutant % ------------------------------------------------- % KAPAR ---- ---- ---- ---- % KAPER ---- ---- ---- ---- % KBPAR ---- ---- ---- ---- % KBPER ---- ---- ---- ---- % KNOR ---- ---- ---- ---- % POLARISER 0.1 ---- ---- ---- % STRAINRET ---- ---- ---- ---- % ARREST ---- ---- ---- ---- % ID_PLUSORG ---- ---- ---- ---- % ID_MINUSORG ---- ---- ---- ---- % V_KAREAL ---- ---- ---- ---- % ID_TIP ---- ---- ---- ---- % ID_TOP ---- ---- ---- ---- % S_GROWTH 0.01 ---- ---- ---- % ID_MID ---- ---- ---- ---- %%% USER CODE: MORPHOGEN INTERACTIONS % In this section you may modify the mesh in any way that does not Section 4 % alter the set of nodes. % Use the same pattern for both submodels RangeTip=(m.nodes(:,1)<-0.8)&... (abs(m.nodes(:,2))<0.2); RangeMid=(m.nodes(:,1)<=0.5)&... (m.nodes(:,1)>-0.5)&... (abs(m.nodes(:,2))<0.3); RangeTops=(m.nodes(:,1)<=max(m.nodes(:,1))&... (m.nodes(:,1)>0.5)&... (abs(m.nodes(:,2))>0.3)); if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. switch modelname case 'PolariserBased' % % One way to set up a morphogen gradient is by ... % Setting up a gradient by clamping the ends (execute only once) P(RangeTip)=0; P(RangeMid)=0.5; P(RangeTops)=1; id_plusorg_p=P; id_minusorg_p(RangeTip)=1; m.morphogenclamp( RangeTops|RangeTip|RangeMid, polariser_i ) = 1; m = leaf_mgen_conductivity( m, 'POLARISER', 0.1 ); %specifies the diffusion rate of polariser m = leaf_mgen_absorption( m, 'POLARISER', 0.0 ); % specifies degradation rate of polariser case 'DifferentialGrowthBased' % P(:)=0; % One way to set up a morphogen gradient is by ... % Setting up a gradient by clamping the ends (execute only once) s_growth_p(RangeTip)=1; s_growth_p(RangeMid)=0.05; s_growth_p(RangeTops)=0.8; m.morphogenclamp( RangeTops|RangeTip|RangeMid, s_growth_i ) = 1; m = leaf_mgen_conductivity( m, 's_growth', 0.001 ); %specifies the diffusion rate of polariser m = leaf_mgen_absorption( m, 's_growth', 0.0 ); % specifies degradation rate of polariser end end BasicGrowth=0.01; switch modelname case 'PolariserBased' % % Every equation to be formatted should end with an at-at Eqn N comment. kapar_p(:) = BasicGrowth; % when isotropic this will be 0.005 kaper_p(:) = 0.0; % when isotropic this will be 0.005 kbpar_p(:) = BasicGrowth; % when isotropic this will be 0.005 kbper_p(:) = 0.0; % when isotropic this will be 0.005 knor_p(:) = 0; % thickness case 'DifferentialGrowthBased' % % Every equation to be formatted should end with an at-at Eqn N comment. kapar_p(:) = BasicGrowth*s_growth_p; %0.01; % when isotropic this will be 0.005 kaper_p(:) = BasicGrowth*s_growth_p; %0.0; % when isotropic this will be 0.005 kbpar_p(:) = BasicGrowth*s_growth_p; %0.01; % when isotropic this will be 0.005 kbper_p(:) = BasicGrowth*s_growth_p; %0.0; % when isotropic this will be 0.005 knor_p(:) = 0; % thickness end v_kareal_p=kapar_p+kaper_p; % total specified areal growth Section 5 %%% END OF USER CODE: MORPHOGEN INTERACTIONS %%% SECTION 3: INSTALLING MODIFIED VALUES BACK INTO MESH STRUCTURE %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. m.morphogens(:,polariser_i) = P; m.morphogens(:,kapar_i) = kapar_p; m.morphogens(:,kaper_i) = kaper_p; m.morphogens(:,kbpar_i) = kbpar_p; m.morphogens(:,kbper_i) = kbper_p; m.morphogens(:,knor_i) = knor_p; m.morphogens(:,strainret_i) = strainret_p; m.morphogens(:,arrest_i) = arrest_p; m.morphogens(:,id_plusorg_i) = id_plusorg_p; m.morphogens(:,id_minusorg_i) = id_minusorg_p; m.morphogens(:,v_kareal_i) = v_kareal_p; m.morphogens(:,id_tip_i) = id_tip_p; m.morphogens(:,id_top_i) = id_top_p; m.morphogens(:,s_growth_i) = s_growth_p; m.morphogens(:,id_mid_i) = id_mid_p; %%% USER CODE: FINALISATION %%% END OF USER CODE: FINALISATION end %%% USER CODE: SUBFUNCTIONS Section 6 function m = local_setproperties( m ) end