AAMToolbox Details: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Software# | [[Software#Analysing_shapes_in_2D_and_3D:_AAMToolbox|Back to Software]] | ||
== | ==<span style="color:Navy;">Shape modelling: what is the AAMToolbox and why'?</span>== | ||
'''We wish to understand''' how biological organs grow to particular shapes. For this we need a tool to help us think through what we expect to see (''GFtbox'') and we need to make measurements of real biological organs to test our expectations (hypotheses). | |||
<br><br> | |||
However, the shapes of biological organs rarely make measurement simple - how do you measure the two or three dimensional (2 or 3D) shape of an ear, leaf or Snapdragon flower? It is not enough to, for example, measure the length and width of a leaf. Why not? | |||
#Length and width are highly correlated and so you really need only one of them | |||
However, the shapes of biological organs rarely make measurement simple - how do you measure the three dimensional (3D) shape of an ear, leaf or Snapdragon flower? | #Length and width do not capture curvature of the edges | ||
We do it by | |||
*digitising the outlines using, for example, ''VolViewer'' | |||
*averaging the shapes of many examples ('''Procrustes''') then find the '''principle components''' that contribute to variations from the mean shape. The different components are linearly independent of each other (not correlated). Typically most of the variation from the mean for simple leaves is captured in just the two principle components. The whole process including projections into scale space is available in the ''AAMToolbox''. | |||
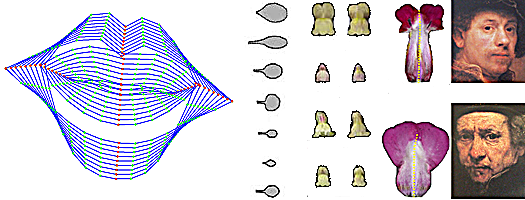
[[image:Various shapes.png|400px|center|Shape and appearance models]]Left - '''lip outlines''' vary along the first principle component. Next - '''leaf and petal''' shapes. Right - Rembrandt's '''self portraits''' vary. | |||
Latest revision as of 15:08, 28 November 2013
Shape modelling: what is the AAMToolbox and why'?
We wish to understand how biological organs grow to particular shapes. For this we need a tool to help us think through what we expect to see (GFtbox) and we need to make measurements of real biological organs to test our expectations (hypotheses).
However, the shapes of biological organs rarely make measurement simple - how do you measure the two or three dimensional (2 or 3D) shape of an ear, leaf or Snapdragon flower? It is not enough to, for example, measure the length and width of a leaf. Why not?
- Length and width are highly correlated and so you really need only one of them
- Length and width do not capture curvature of the edges
We do it by
- digitising the outlines using, for example, VolViewer
- averaging the shapes of many examples (Procrustes) then find the principle components that contribute to variations from the mean shape. The different components are linearly independent of each other (not correlated). Typically most of the variation from the mean for simple leaves is captured in just the two principle components. The whole process including projections into scale space is available in the AAMToolbox.
Left - lip outlines vary along the first principle component. Next - leaf and petal shapes. Right - Rembrandt's self portraits vary.