BioformatsConverter: Difference between revisions
JeromeAvondo (talk | contribs) mNo edit summary |
JeromeAvondo (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
'''DOWNLOAD:''' [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/bioformatsConverter_rev2480.zip bioformatsConverter_rev2480.zip] | '''DOWNLOAD:''' [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/bioformatsConverter_rev2480.zip bioformatsConverter_rev2480.zip] | ||
Revision as of 17:08, 6 May 2010
DOWNLOAD: bioformatsConverter_rev2480.zip
Documentation
This software is useful for batch converting microscopy image formats, like STK, LSM, LIE, ZVI etc etc, to OME-TIFF or PNG files. See this page for the supported image formats. The advantage of converting to OME-TIFF over regular TIFF files is that most of the original image metadata is kept in the file.
This tool is based on the Bioformats Library developed by the University of Wisconsin-Madison.
Windows Installation
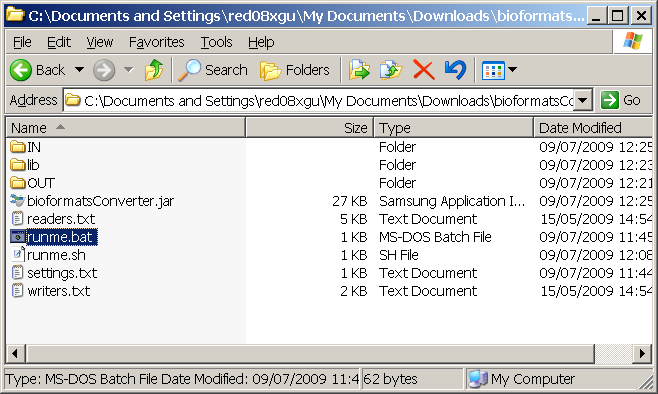
Extract the contents of the zip file to somewhere sensible on your local machine. What you should end up with is the following:
Then you can place your image files in the IN/ folder. Note you can place files of different image file formats. ie: a mix of LSM, STK etc...
You then double click on the runme.bat and wait for the program to finish.
This will save your converted images in the OUT/ directory.
Configuration
The program is configured by editing the file called Settings.txt. This file can be edited by double clicking on it.
You have the following options.
savemode: TIFF
With this option you can either tell it to save as TIFF or PNG files.
The second option is only relevant if you are saving as TIFF files. If you are saving as PNG this option has no effect.
stackmode: Z_STACK_TIFFS
This option can take the following options: SINGLE_TIFFS, Z_STACK_TIFFS, ZC_STACK_TIFFS or ZCT_STACK_TIFFS
SINGLE_TIFFS: this splits the z-series, channels and timepoints as individual tiff files
Z_STACK_TIFFS: this splits the channels and timepoints as individual z-series tiff stack files
ZC_STACK_TIFFS: this splits the timepoints as individual z-series and multichannels tiff stack files
ZCT_STACK_TIFFS: this creates a single tiff stack for the z-series, mutlichannels and timepoints