Measuring using interaction function subroutines
Jump to navigation
Jump to search
Measuring growth of a model
Download a project illustrating measurements GPT_MeasureGrowth_20121203
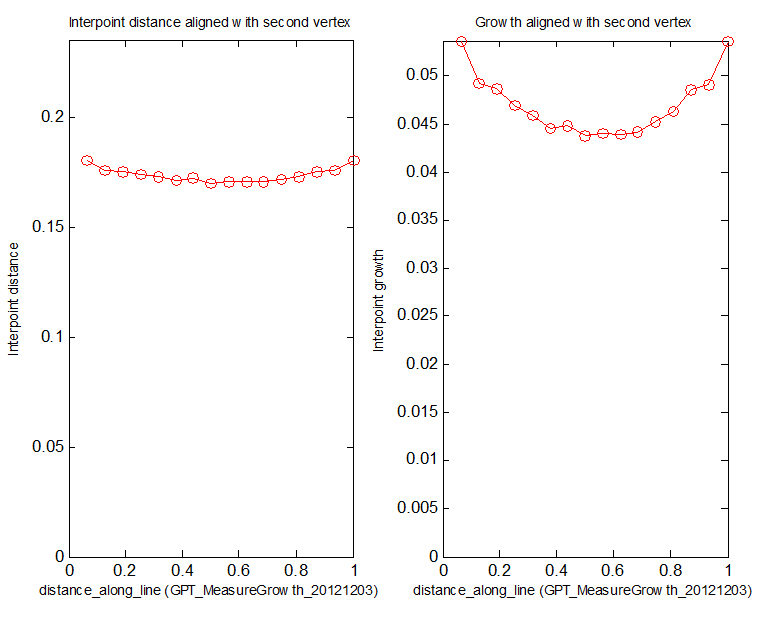
The GPT_MeasureGrowth_20121203 interaction function is shown at the bottom of this page.
There are several ways to measure growth on the model. For example, there are two auxiliary functions:
[out1, out2, out3, out4] = leaf_computeGrowthFromDisplacements( m, displacements, time, ... ) Compute the growth rate in each FE of m given the displacements of all the prism nodes and the time over which these displacements happened. This assumes that m is in the state after these displacements have happened. The results returned depend on the options and on how many output arguments are given. Note that unlike most leaf_* commands, m itself is not one of the output arguments, since this procedure does not need to modify m.
function m=leaf_profile_monitor(m,realtime,RegionLabels,Morphogens,start_figno)
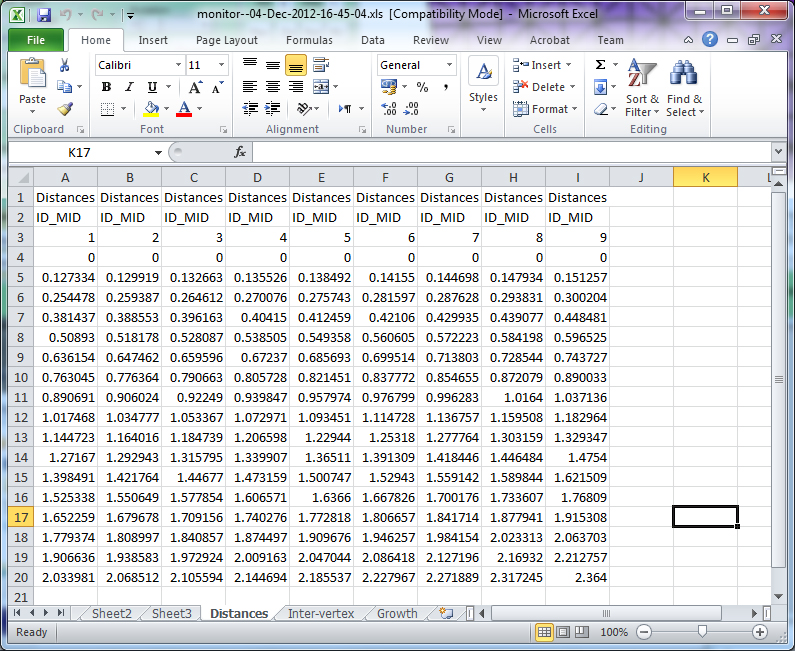
monitor morphogen levels at a set of vertices
m, mesh
RegionLabels, vertices to be monitored as designated by cell array of strings, i.e. region labels
Morphogens, cell array of strings, i.e. uppercase morphogen names to
be monitored. There should be one RegionLabels string for each
Morphogens string
Vertlabels, if true then display vertex numbers in each region label on
the mesh default false
start_figno, default figure 1 (Must open a fig even if just monitoring to file)
MonData, optional output of data structure
- Measuring growth
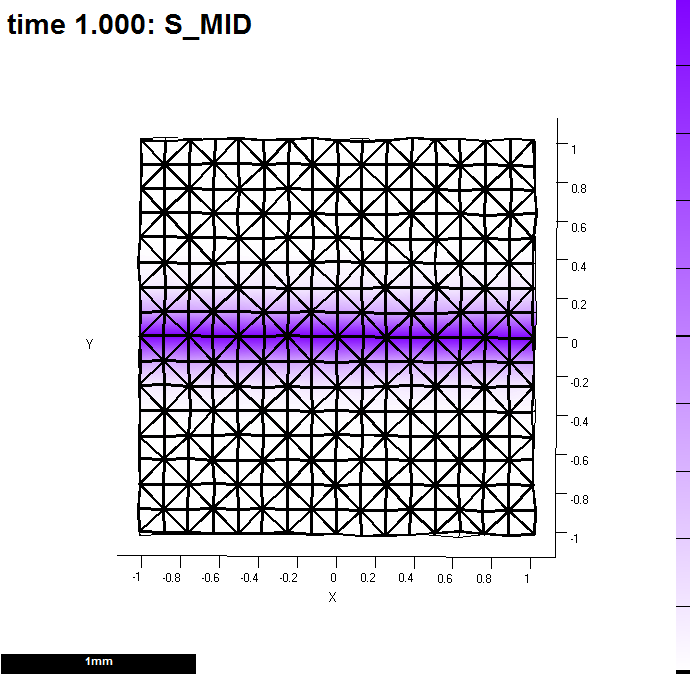
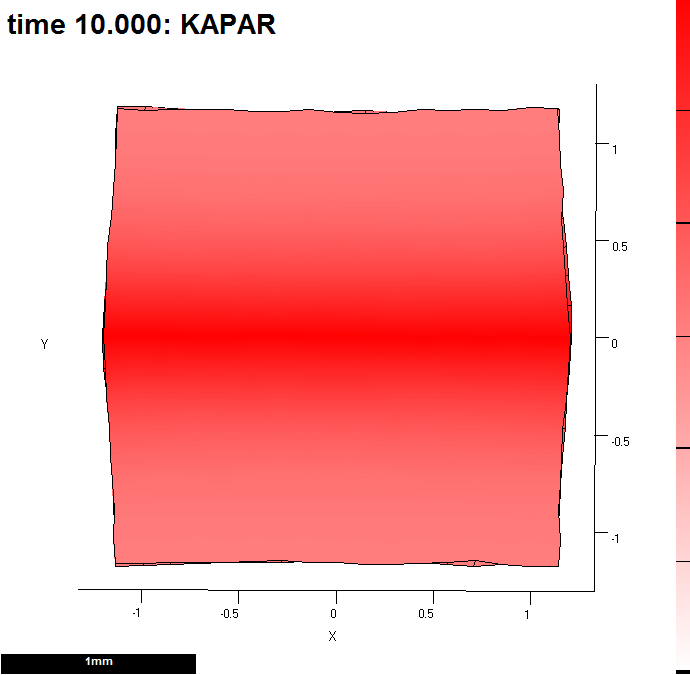
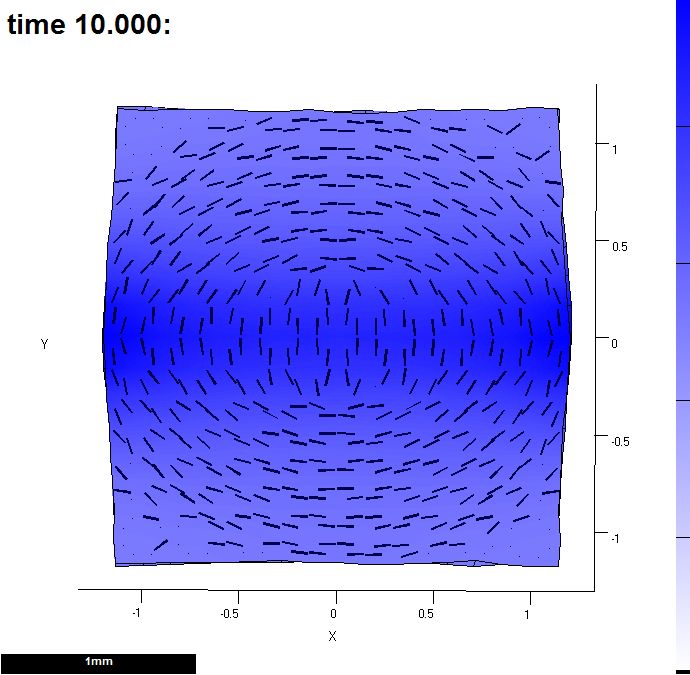
function m = gpt_measuregrowth_20121203( m ) %m = gpt_measuregrowth_20121203( m ) % Morphogen interaction function. % Written at 2012-12-04 15:01:27. % GFtbox revision 4418, . % The user may edit any part of this function between delimiters % of the form "USER CODE..." and "END OF USER CODE...". The % delimiters themselves must not be moved, edited, deleted, or added. if isempty(m), return; end fprintf( 1, '%s found in %s\n', mfilename(), which(mfilename()) ); try m = local_setproperties( m ); catch end realtime = m.globalDynamicProps.currenttime; %%% USER CODE: INITIALISATION %%% END OF USER CODE: INITIALISATION %%% SECTION 1: ACCESSING MORPHOGENS AND TIME. %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. if isempty(m), return; end setGlobals(); global gNEW_KA_PAR gNEW_KA_PER gNEW_KB_PAR gNEW_KB_PER global gNEW_K_NOR gNEW_POLARISER gNEW_STRAINRET gNEW_ARREST dt = m.globalProps.timestep; polariser_i = gNEW_POLARISER; P = m.morphogens(:,polariser_i); [kapar_i,kapar_p,kapar_a,kapar_l] = getMgenLevels( m, 'KAPAR' ); [kaper_i,kaper_p,kaper_a,kaper_l] = getMgenLevels( m, 'KAPER' ); [kbpar_i,kbpar_p,kbpar_a,kbpar_l] = getMgenLevels( m, 'KBPAR' ); [kbper_i,kbper_p,kbper_a,kbper_l] = getMgenLevels( m, 'KBPER' ); [knor_i,knor_p,knor_a,knor_l] = getMgenLevels( m, 'KNOR' ); [strainret_i,strainret_p,strainret_a,strainret_l] = getMgenLevels( m, 'STRAINRET' ); [arrest_i,arrest_p,arrest_a,arrest_l] = getMgenLevels( m, 'ARREST' ); [id_mid_i,id_mid_p,id_mid_a,id_mid_l] = getMgenLevels( m, 'ID_MID' ); [s_mid_i,s_mid_p,s_mid_a,s_mid_l] = getMgenLevels( m, 'S_MID' ); % Mesh type: rectangle % base: 0 % centre: 0 % randomness: 0.1 % version: 1 % xdivs: 16 % xwidth: 2 % ydivs: 16 % ywidth: 2 % Morphogen Diffusion Decay Dilution Mutant % -------------------------------------------------- % KAPAR ---- ---- ---- ---- % KAPER ---- ---- ---- ---- % KBPAR ---- ---- ---- ---- % KBPER ---- ---- ---- ---- % KNOR ---- ---- ---- ---- % POLARISER ---- ---- ---- ---- % STRAINRET ---- ---- ---- ---- % ARREST ---- ---- ---- ---- % ID_MID ---- ---- ---- ---- % S_MID 0.01 ---- ---- ---- %%% USER CODE: MORPHOGEN INTERACTIONS if (Steps(m)==0) && m.globalDynamicProps.doinit % Initialisation code. % assign a morphogen down the middle of the y axis id_mid_p((m.nodes(:,2)<0.1)&(m.nodes(:,2)>-0.1))=1; s_mid_p(:)=0; s_mid_p=id_mid_p; m.morphogenclamp( (id_mid_p==1), s_mid_i ) = 1; m = leaf_mgen_conductivity( m, 's_mid', 0.01 ); %specifies the diffusion rate of polariser m = leaf_mgen_absorption( m, 's_mid', 0.0 ); % specifies degradation rate of polariser end kapar_p=15 * pro(1,s_mid_p); kbpar_p=15 * pro(1,s_mid_p); kaper_p=10 * pro(1,s_mid_p); kbper_p=10 * pro(1,s_mid_p); knor_p =0; % calculate growth over dt interval if isfield(m.userdata,'oldpos') displacements = m.prismnodes - m.userdata.oldpos; [growth,gf] = leaf_computeGrowthFromDisplacements( m, displacements, ... realtime - m.userdata.starttime,'axisorder', 'maxminnor', ... 'anisotropythreshold', 0.05); % plot resultant areal growth rates over dt (i.e. one step). m = leaf_plotoptions( m, 'pervertex',perFEtoperVertex(m,sum(growth(:,1:2),2)) ,'perelementaxes', gf(:,1,:), 'drawtensoraxes', true ); % monitor properties of vertices must be done here - so that it reports newly equilibrated levels end m.userdata.oldpos = m.prismnodes; m.userdata.starttime = realtime; % monitor growth in a Figure and write excel file [m,MonData]=leaf_profile_monitor(m,... % essential 'REGIONLABELS',{'ID_MID'},... % essential 'VERTLABELS',false,'FigNum',1,'EXCEL',true); % optional (file in snapshots directory') % if sum(ismember(output,realtime)) && m.userdata.output ==1 % % Output - plot an image at high resolution % path = fileparts(which(m.globalProps.modelname)); % [m,ok] = leaf_snapshot( m,[path,filesep,'snapshots',filesep,modelname,'_',modeltype,'.png'], 'resolution',[]); % % % end %%% END OF USER CODE: MORPHOGEN INTERACTIONS %%% SECTION 3: INSTALLING MODIFIED VALUES BACK INTO MESH STRUCTURE %%% AUTOMATICALLY GENERATED CODE: DO NOT EDIT. m.morphogens(:,polariser_i) = P; m.morphogens(:,kapar_i) = kapar_p; m.morphogens(:,kaper_i) = kaper_p; m.morphogens(:,kbpar_i) = kbpar_p; m.morphogens(:,kbper_i) = kbper_p; m.morphogens(:,knor_i) = knor_p; m.morphogens(:,strainret_i) = strainret_p; m.morphogens(:,arrest_i) = arrest_p; m.morphogens(:,id_mid_i) = id_mid_p; m.morphogens(:,s_mid_i) = s_mid_p; %%% USER CODE: FINALISATION %%% END OF USER CODE: FINALISATION end