Types of morphogens and factors: Difference between revisions
No edit summary |
No edit summary |
||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | |||
[[GFtbox Tutorial pages| Back to Tutorial pages]]<br> | |||
[[Ready Reference Manual|Ready Reference Manual]]<br><br> | |||
===Notes on Matlab syntax=== | |||
*Scalar: a single value | |||
*Vector: a column of values - one value of each node (vertex) in the mesh. | |||
In an interaction function a variable, lets use a variable called ''id_a_p''. This is a vector with one element for each node (vertex). | |||
*''id_a_p(:)=0''; | |||
sets the value of all the nodes to 0 ( the colon : means all). | |||
*''id_a_p(1)=3''; | |||
sets the value of node 1 to 3. Actually, we rarely know where the nodes are - and we don't care. If we want to set up a pattern of nodes we usually refer to them by their spatial position. Thus, we could find all nodes that have an ''x'' value that is equal to the minimum ''x'' value in the mesh. | |||
*''minx=min(m.nodes(:,1));'' | |||
sets ''minx'' to the minimum ''x'' and the expression'' (m.nodes(:,1)==minx)'' finds all nodes with an ''x'' value that equals the minimum ''x'' value. Thus, | |||
*''id_a_p(m.nodes(:,1)==minx)=3;'' | |||
sets ''id_a_p'' at the selected nodes to the value 3. | |||
<br><br> | |||
('''Note to programmers''': we could have produced a '''class library''' to handle the different types of variable - identity factors, signals, etc. However, in our view it is too early in the development of software for modelling the growth of biological tissues to be confident of what will be required. Too build a class library now would be to build a '''conceptual straightjacket that would inhibit further research'''.)<br><br> | |||
=''GFtbox'' Growth factor naming conventions= | =''GFtbox'' Growth factor naming conventions= | ||
| Line 6: | Line 21: | ||
{| border="0" cellpadding="1" cellspacing="1" | {| border="0" cellpadding="1" cellspacing="1" | ||
|- valign="top" | |- valign="top" | ||
| width="30"| id_ || identity factors (non-diffusible). Identity factors can be labelled as on the A or B side (ida_, idb_) | | width="30"| id_ || '''identity''' factors (non-diffusible). Identity factors can be labelled as on the A or B side (ida_, idb_) | ||
|- valign="top" | |- valign="top" | ||
| width="30"| s_ || signalling factors (diffuse | | width="30"| s_ || '''signalling''' factors (diffuse). Polariser level is unique and has a particular name ''P''. | ||
|- | |- | ||
| width="30"| v_ || visualising labels (e.g. theta, bend, kpar, other labels) | | width="30"| v_ || visualising labels (e.g. theta, bend, kpar, other labels) | ||
| Line 17: | Line 32: | ||
{| border="0" cellpadding="1" cellspacing="1" | {| border="0" cellpadding="1" cellspacing="1" | ||
|- valign="top" | |- valign="top" | ||
| width="30"| _p || promotor: how much the expression of the factor is being promoted in the model | | width="30"| _p || vector representing '''promotor''': how much the expression of the factor is being promoted in the model at every node (vertex) in the mesh. | ||
|- valign="top" | |- valign="top" | ||
| width="30"| _l || level of morphogen, i.e. *_p times *_a. This gets updated at the end of the interaction function and might change according to growth or mutation, see *_a. | | width="30"| _l || vector representing the '''level''' of morphogen, i.e. *_p times *_a at every node in the mesh. This gets updated at the end of the interaction function and might change according to growth or mutation, see *_a. | ||
|- valign="top" | |- valign="top" | ||
| width="30"| _i || index into the main data structure morphogen field, m. | | width="30"| _i || scalar index into the main data structure morphogen field, m.morphogens. | ||
|- valign="top" | |- valign="top" | ||
| width="30"| _a || activity: by default it is 1 and if the factor is mutated or silenced then it is 0. However, it can be in the range [0,...,1]. | | width="30"| _a || scalar specifying activity: by default it is 1 and if the factor is mutated or silenced then it is 0. However, it can be in the range [0,...,1]. | ||
|} | |} | ||
=Standard morphogens= | =Standard morphogens= | ||
| Line 29: | Line 44: | ||
{| border="0" cellpadding="1" cellspacing="1" | {| border="0" cellpadding="1" cellspacing="1" | ||
|- valign="top" | |- valign="top" | ||
| width="200"| kapar_p,kapar_a,kapar_l || | | width="200"|'' '''kapar_p''', kapar_a, kapar_l'' || vector that specifies growth on the 'A' side '''parallel''' to the polariser and in the plane of the tissue at every node. | ||
|- valign="top" | |- valign="top" | ||
| width="200"| kbpar_p,kbpar_a,kbpar_l || | | width="200"| '''''kbpar_p''', kbpar_a, kbpar_l'' || vector that specifies growth on the 'B' side parallel to the polariser and in the plane of the tissue at every node. | ||
|- valign="top" | |- valign="top" | ||
| width="200"| kaper_p,kaper_a,kaper_l || | | width="200"| '''''kaper_p''', kaper_a, kaper_l'' || vector that specifies growth on the 'A' side '''perpendicular''' to the polariser and in the plane of the tissue at every node. | ||
|- valign="top" | |- valign="top" | ||
| width="200"| kbper_p,kbper_a,kbper_l || | | width="200"| '''''kbper_p''', kbper_a, kbper_l'' || vector that specifies growth on the 'B' side perpendicular to the polariser and in the plane of the tissue at every node. | ||
|- valign="top" | |- valign="top" | ||
| width="200"| knor_p,knor_a,knor_l | | width="200"| '''''knor_p''', knor_a, knor_l'' || vector that specifies growth '''normal''' to the plane of the tissue (i.e. thickness) at every node. | ||
|} | |} | ||
==Controlling growth== | ==Controlling growth== | ||
{| border="0" cellpadding="1" cellspacing="1" | {| border="0" cellpadding="1" cellspacing="1" | ||
|- valign="top" | |- valign="top" | ||
| width="200"| P || Polariser level. | | width="200"| '''''P''''' || vector that specifies '''Polariser level''' at every node. | ||
|- valign="top" | |- valign="top" | ||
| width="200"| strainret_p,strainret_a,strainret_l || a number between 0 (no strain retention, default) and 1 (full strain retention). | | width="200"| ''strainret_p, strainret_a, strainret_l'' || vector that specifies strain retention at every node. It is a number between 0 (no strain retention, default) and 1 (full strain retention). | ||
|- valign="top" | |- valign="top" | ||
| width="200"| | | width="200"| ''arrest_p,arrest_a,arrest_l'' || vector that specifies an overall inhibition of growth at every node of the mesh. It is a number between 0 (no overall inhibition of growth, default) and 1 (fully inhibited). | ||
|} | |||
=User (your) morphogens= | |||
Within the interaction function all the usual Matlab variables and types are available. Variables that are to be visible from the GUI must follow certain conventions and be declared within the GUI, see below. | |||
==Identity factors, signalling factors, viewable variables== | |||
Growth is all about patterns of growth factors interacting in space (spatially) and over time (temporally). It is, therefore, essential to be able to view them on the mesh. This is the main purpose of the graphical user interface (GUI). Such variables are created (declared) from the GUI as follows. They have a name e.g. ''id_plusorg''. This is how the variable will be known as a case insensitive string (according to Matlab conventions, 'id_plusorg' or 'ID_PLUSORG'). Within the interaction function this name will be lowercase and have a suffix e.g. ''id_plusorg_p'' - see above. | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
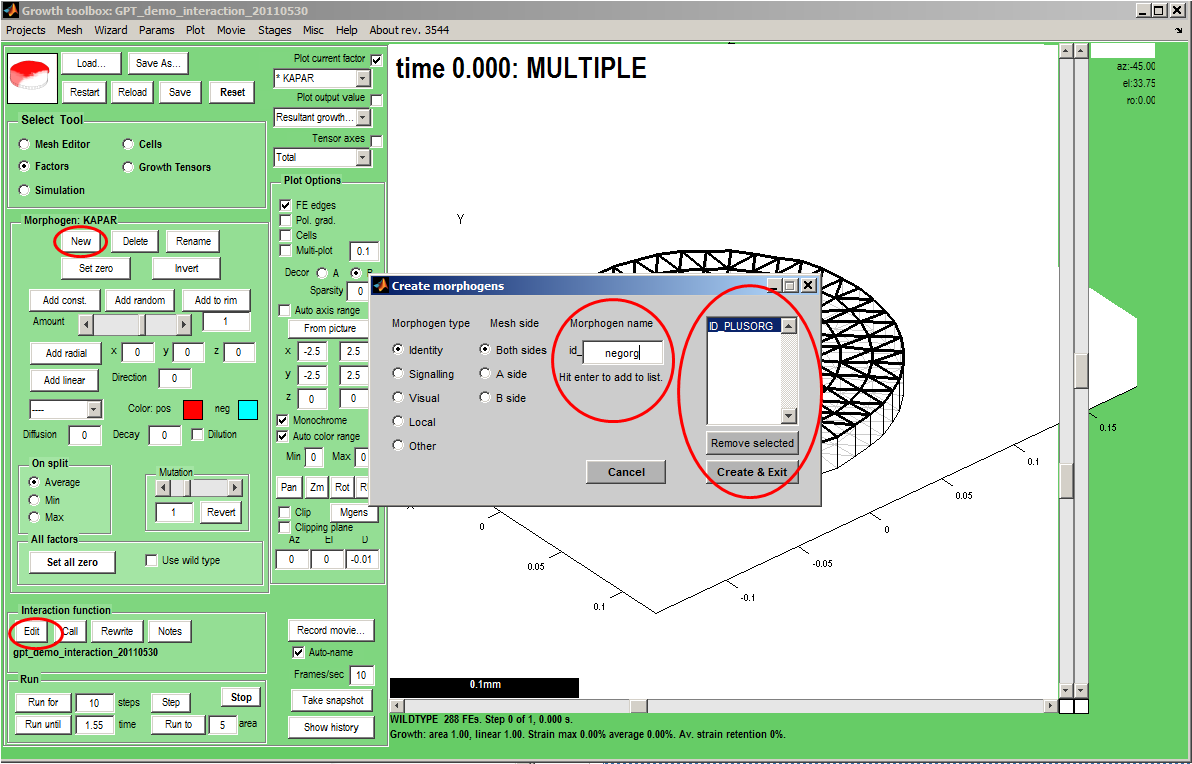
|width="300px"|'''Adding (declaring) variables''' that are visible in the GUI. ''Select Tool:Factors'' makes the ''Morphogen'' Panel visible. We add a two new morphogens, ''id_plusorg'' and ''id_negorg''. Select a type (in this case ''id_''), key in the name, press Enter and the name will be added to a list. Finally, create the morphogens and exit the dialogue. Select a colour for each, ''Panel: Morphogen:Color. pos.'' and ''Color. neg'' (for negative values).<br><br> '''Save''' to make sure they are saved in the Mesh. Pressing ''Panel:Interaction Function:Edit'' '''will reload the editor''' with a revised copy of the interaction function. The new copy will retain any edits that you have made already together with the new variables. | |||
|width="500px"|[[File:AddingMorphogens.png|500px|GFtbox interface adding morphogens]] | |||
|} | |} | ||
Latest revision as of 10:27, 15 December 2012
Back to Tutorial pages
Ready Reference Manual
Notes on Matlab syntax
- Scalar: a single value
- Vector: a column of values - one value of each node (vertex) in the mesh.
In an interaction function a variable, lets use a variable called id_a_p. This is a vector with one element for each node (vertex).
- id_a_p(:)=0;
sets the value of all the nodes to 0 ( the colon : means all).
- id_a_p(1)=3;
sets the value of node 1 to 3. Actually, we rarely know where the nodes are - and we don't care. If we want to set up a pattern of nodes we usually refer to them by their spatial position. Thus, we could find all nodes that have an x value that is equal to the minimum x value in the mesh.
- minx=min(m.nodes(:,1));
sets minx to the minimum x and the expression (m.nodes(:,1)==minx) finds all nodes with an x value that equals the minimum x value. Thus,
- id_a_p(m.nodes(:,1)==minx)=3;
sets id_a_p at the selected nodes to the value 3.
(Note to programmers: we could have produced a class library to handle the different types of variable - identity factors, signals, etc. However, in our view it is too early in the development of software for modelling the growth of biological tissues to be confident of what will be required. Too build a class library now would be to build a conceptual straightjacket that would inhibit further research.)
GFtbox Growth factor naming conventions
Prefix
| id_ | identity factors (non-diffusible). Identity factors can be labelled as on the A or B side (ida_, idb_) |
| s_ | signalling factors (diffuse). Polariser level is unique and has a particular name P. |
| v_ | visualising labels (e.g. theta, bend, kpar, other labels) |
| f_ | finite element manipulation (e.g. seams, making holes) |
Suffix
| _p | vector representing promotor: how much the expression of the factor is being promoted in the model at every node (vertex) in the mesh. |
| _l | vector representing the level of morphogen, i.e. *_p times *_a at every node in the mesh. This gets updated at the end of the interaction function and might change according to growth or mutation, see *_a. |
| _i | scalar index into the main data structure morphogen field, m.morphogens. |
| _a | scalar specifying activity: by default it is 1 and if the factor is mutated or silenced then it is 0. However, it can be in the range [0,...,1]. |
Standard morphogens
Specifying growth
| kapar_p, kapar_a, kapar_l | vector that specifies growth on the 'A' side parallel to the polariser and in the plane of the tissue at every node. |
| kbpar_p, kbpar_a, kbpar_l | vector that specifies growth on the 'B' side parallel to the polariser and in the plane of the tissue at every node. |
| kaper_p, kaper_a, kaper_l | vector that specifies growth on the 'A' side perpendicular to the polariser and in the plane of the tissue at every node. |
| kbper_p, kbper_a, kbper_l | vector that specifies growth on the 'B' side perpendicular to the polariser and in the plane of the tissue at every node. |
| knor_p, knor_a, knor_l | vector that specifies growth normal to the plane of the tissue (i.e. thickness) at every node. |
Controlling growth
| P | vector that specifies Polariser level at every node. |
| strainret_p, strainret_a, strainret_l | vector that specifies strain retention at every node. It is a number between 0 (no strain retention, default) and 1 (full strain retention). |
| arrest_p,arrest_a,arrest_l | vector that specifies an overall inhibition of growth at every node of the mesh. It is a number between 0 (no overall inhibition of growth, default) and 1 (fully inhibited). |
User (your) morphogens
Within the interaction function all the usual Matlab variables and types are available. Variables that are to be visible from the GUI must follow certain conventions and be declared within the GUI, see below.
Identity factors, signalling factors, viewable variables
Growth is all about patterns of growth factors interacting in space (spatially) and over time (temporally). It is, therefore, essential to be able to view them on the mesh. This is the main purpose of the graphical user interface (GUI). Such variables are created (declared) from the GUI as follows. They have a name e.g. id_plusorg. This is how the variable will be known as a case insensitive string (according to Matlab conventions, 'id_plusorg' or 'ID_PLUSORG'). Within the interaction function this name will be lowercase and have a suffix e.g. id_plusorg_p - see above.