MTtbox documentation: Difference between revisions
Jump to navigation
Jump to search
(Created page with 'Return to Bangham Lab Software<br><br> ==Current Status== MTtbox is currently under test and further development<br> The main data structure is called: 'data'…') |
No edit summary |
||
| Line 20: | Line 20: | ||
{| border="0" cellpadding="5" cellspacing="3" | {| border="0" cellpadding="5" cellspacing="3" | ||
|- valign="top" | |- valign="top" | ||
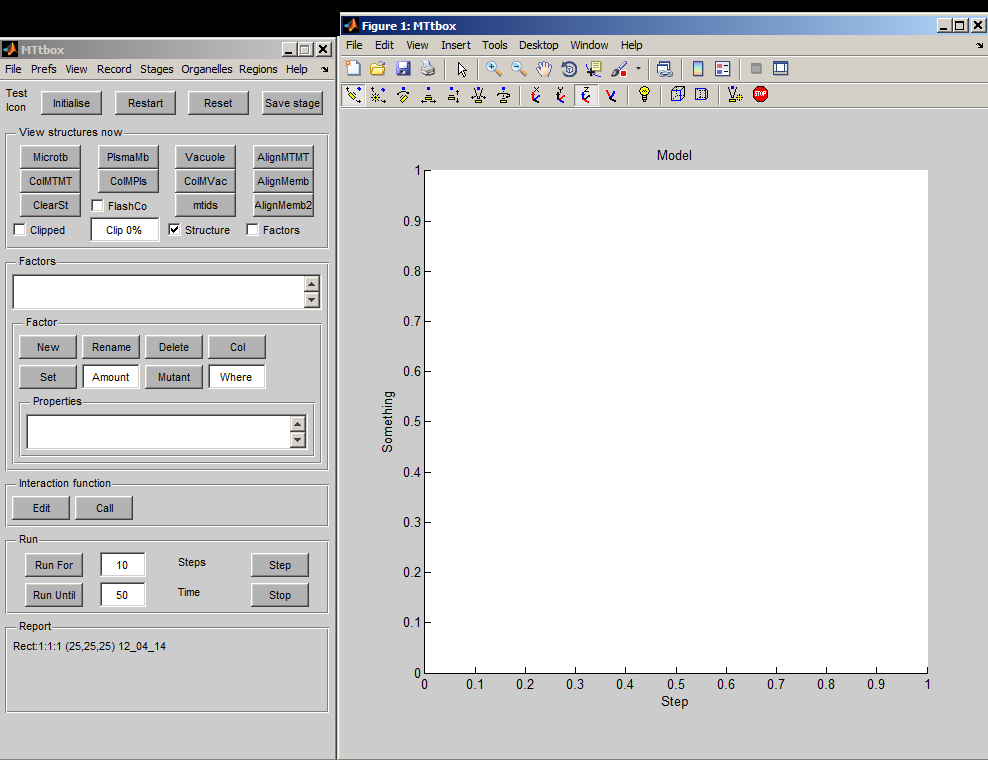
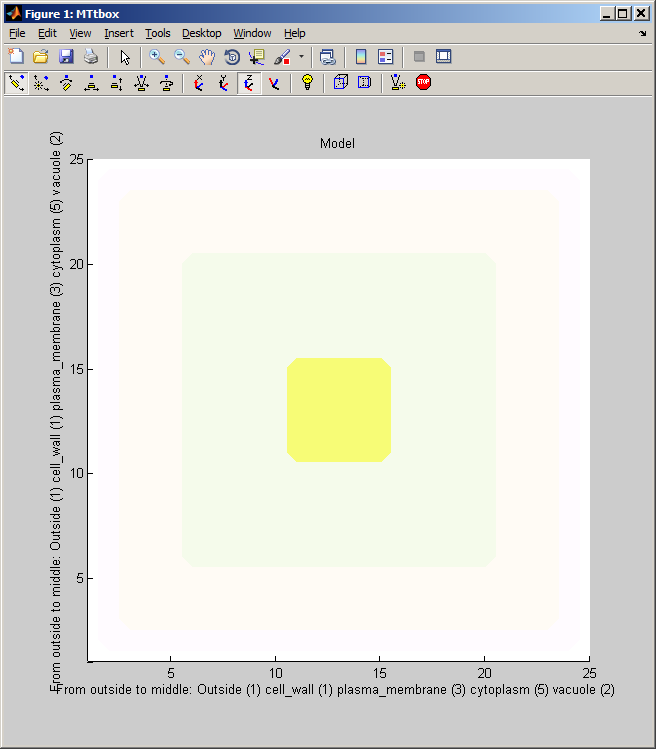
|width="500pt"|A default project is created by selecting: menu:File:New Project<br><br>It forms a cell bounded by regions labelled: Outside, cell_wall, plasma_membrane, cytoplasm and vacuole. These are concentric volumes. Length is measured in microns (time in seconds). The axis labels indicate the thickness of each layer in terms of voxels in this particular model. This is extremely low resolution - it means that the microtubules will have to be unrealistically large. This is convenient for debugging and illustrating the system but not for simulations. The outer surface of each region is coloured, e.g. the vacuole is yellow and the cytoplasm is green.<br><br> | |width="500pt"|A default project is created by selecting: ''menu:File:New Project''<br><br>It forms a cell bounded by regions labelled: Outside, cell_wall, plasma_membrane, cytoplasm and vacuole. These are concentric volumes. Length is measured in microns (time in seconds). The axis labels indicate the thickness of each layer in terms of voxels in this particular model. This is extremely low resolution - it means that the microtubules will have to be unrealistically large. This is convenient for debugging and illustrating the system but not for simulations. The outer surface of each region is coloured, e.g. (Fig. on right) the vacuole is yellow and the cytoplasm is pale green.<br><br> | ||
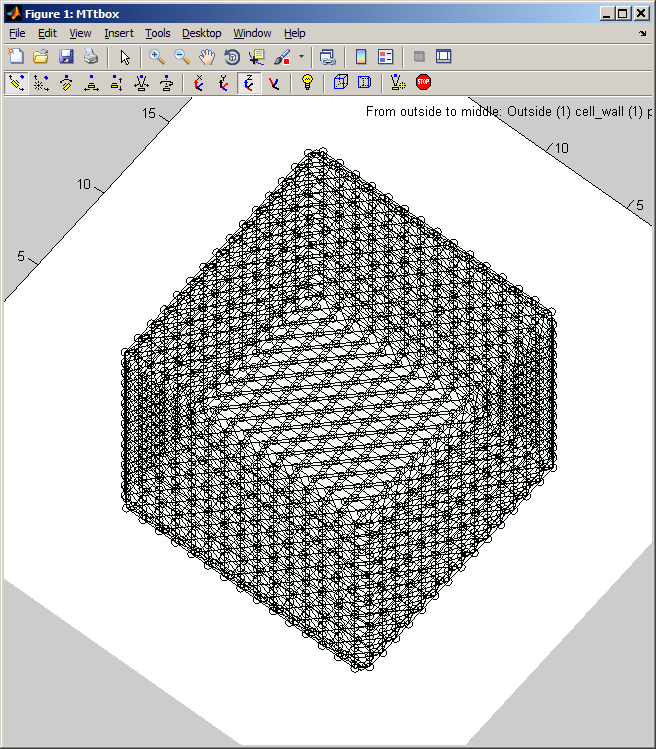
The cell can be rotated etc. using the panels at the top of the display panel. | The cell can be rotated etc. using the panels at the top of the display panel. Fig. below: all the regions have been hidden (uncheck each item in menu:View) and the mesh associated with the cytoplasm outer surface is displayed (check ''menu:View:Organelle meshes'')<br> | ||
[[Image:MTtboxCytoplasmMesh.png|300px|MTtbox GUI]] | [[Image:MTtboxCytoplasmMesh.png|300px|MTtbox GUI]] | ||
|width="400pt"|[[Image:MTtboxDefaultProject.png|400px|MTtbox GUI]] | |||
|} | |||
===1 C=== | |||
{| border="0" cellpadding="5" cellspacing="3" | |||
|- valign="top" | |||
|width="500pt"|A project is saved by selecting: ''menu:File:Save as''<br><br>Having first saved a project a default Interaction Function is created by selecting ''Edit''. A default project file [[MTtbox default interaction function|(example of the default interaction function)]] contains lots of comments to provide help on how to develop the project.<br><br>At present the Interaction Functions is not copied to the new project on each Save as command - this has to be done manually. | |||
|width="400pt"|[[Image:MTtboxDefaultProject.png|400px|MTtbox GUI]] | |width="400pt"|[[Image:MTtboxDefaultProject.png|400px|MTtbox GUI]] | ||
|} | |} | ||
Revision as of 17:02, 27 April 2012
Return to Bangham Lab Software
Current Status
MTtbox is currently under test and further development
The main data structure is called: 'data'. It can be accessed from the Matlab command line by declaring data to be global.
global data
at any time. The following documentation will refer to fields in data. It also refers to the custom menu items by menu:name.
Graphical User Interface
1 A
1 B
1 C
| A project is saved by selecting: menu:File:Save as Having first saved a project a default Interaction Function is created by selecting Edit. A default project file (example of the default interaction function) contains lots of comments to provide help on how to develop the project. At present the Interaction Functions is not copied to the new project on each Save as command - this has to be done manually. |
|