Software: Difference between revisions
No edit summary |
Fixing the formatting. |
||
| (255 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | __TOC__ | ||
<!-- ===[[BanghamLabSVN|<span style="color: Navy">Copy of BanghamLab source code</span>]]=== --> | |||
<span style="color: DarkGreen">'''Current activity: a collaboration''' with the [http://rico-coen.jic.ac.uk/index.php/Main_Page CoenLab] with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.</span> | |||
===<span style="color:DarkGreen;">Notes on documenting our software</span>=== | |||
[[Tricks for documenting software|Notes for Lab members on how to contribute to this Wiki and where to put downloads. ]] | |||
<br>Matlab tip: searching a large data structure for a particular field. Clear the command window. Evaluate the structure to list all the fields, then use the usual control-f search tool on the command window. | |||
=<span style="color:DarkGreen;">'''Computational biology'''</span>= | |||
==<span style="color:DarkGreen;">Quantitative understanding of growing shapes: '''GFtbox'''</span>== | |||
====<span style="color:#A52A2A;">We developed ''GFtbox'' to allow us to model the growth of complex shapes with the ultimate goal: to understand the relationships between genes, growth and form.</span>==== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="400"| [[Image:Journal.pbio.1000537.g009.png|350px|Growth of a flower]] Example of a growing snapdragon flower and some mutants ( [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.1000537 Green et al 2011]). Growth is specified by factors (genes) according to the Growing Polarised Tissue Framework. Colours represent putative gene activity, arrows the polariser gradient and spots clones. | |||
|<wikiflv width="300" height="313" logo="false" loop="true" background="white" position="right" autoplay='true'>GPT_Snapdragon_2010_Green_et_al-0002_1.flv|GPT_Snapdragon_2010_Green_et_al-0002.png</wikiflv> | |||
<!--|<wikiflv width="280" height="300" logo="false" loop="true" background="white">Journal.pbio.1000537.s025_1.flv|Journal.pbio.1000537.g009.png</wikiflv>--> | |||
|} | |||
{| border="0" cellpadding="5" cellspacing="5" | {| border="0" cellpadding="5" cellspacing="5" | ||
|- valign="top" | |- valign="top" | ||
| Line 8: | Line 24: | ||
|width="40%"| | |width="40%"| | ||
For modelling the growth of shapes. <br><br> | For modelling the growth of shapes. <br><br> | ||
[[Ready Reference Manual|'''''Ready Reference''''' Manual]]<br><br> | [[Ready Reference Manual|'''''Ready Reference''''' Manual]]<br><br> | ||
(PC, Mac, Linux, uses Matlab<br>no Mathworks toolboxes needed<br>[http://www.mathworks.com/products/matlab/tryit.html Matlab 30 day free trial] and <br>[http://www.mathworks.com/academia/student_version/?s_cid=global_nav student edition])<br><br> | (PC, Mac, Linux, uses Matlab<br>no Mathworks toolboxes needed<br>[http://www.mathworks.com/products/matlab/tryit.html Matlab 30 day free trial] and <br>[http://www.mathworks.com/academia/student_version/?s_cid=global_nav student edition])<br><br> | ||
| | |||
[[Ready Reference Manual ancillary functions|'''''Ready Reference'for ancillary functions: in '''interaction function''' and external functions'''' Manual]]<br><br> | |||
[[GFtbox|'''What? How? Where?''']]<span style="color: Green"> Background</span><br><br> | |||
[[GFtbox Tutorial pages|'''''Tutorials''''': from the beginning]]<span style="color: Green"> Start here</span><br><br> | |||
[[GFtbox Example pages|'''''Examples''''': from publications]]<br><br> | |||
[https://github.com/JIC-Enrico-Coen/GrowthToolbox <span style="color: Gray">'''''Download GFTbox''''' from GitHub</span>] | |||
[https://github.com/JIC-Enrico-Coen <span style="color: Gray">'''''Download GFTbox models''''' from GitHub</span>]<br><br> | |||
|} | |} | ||
==<span style="color:DarkGreen;">VolViewer== | |||
==<span style="color:DarkGreen;">Viewing and measuring volume images: '''VolViewer'''== | |||
{| border="0" cellpadding="5" cellspacing="5" | {| border="0" cellpadding="5" cellspacing="5" | ||
|- valign="top" | |- valign="top" | ||
|width="10%"| <imgicon>VolViewer-logo.png|120px|VolViewer</imgicon> | |width="10%"| <imgicon>VolViewer-logo.png|120px|VolViewer</imgicon> | ||
|width="40%"|For viewing and measuring | |width="40%"|For viewing and measuring '''volume images''' on both normal and '''stereo''' screens. Typical images from: confocal microscope and Optical Projection Tomography (OPT) images<br><br> | ||
(Windows, Mac, Linux) | [[VolViewer#Description|'''What? How? Where?''']]<br><br> | ||
|width="50%"| VolViewer | [[VolViewer#User Documentation|'''''Tutorials''''': from the beginning]]<br><br> | ||
[[VolViewer#Download| '''''Download''''']]<br><br> | |||
(Windows, Mac, Linux)<br><br> | |||
Output from VolViewer has appeared in:<br> | |||
[http://www.cell.com/cell_picture_show-plantbio Cell: Online Gallery] | [http://www.amazon.co.uk/Handbook-Plant-Science-Keith-Roberts/dp/0470057238/ref=sr_1_19?s=books&ie=UTF8&qid=1289321357&sr=1-19 Front cover: Handbook of Plant Science] | [http://www.plantcell.org/content/18/9.toc Front cover: The Plant Cell] | [http://www.americanscientist.org/issues/pub/2013/1/3d-carnivorous-plants American Scientist] | [http://www.rms.org.uk/Resources/Royal%20Microscopical%20Society/infocus/Edgar%20article.pdf Royal Microscopical Society: Infocus Magazine] | [http://www.bioptonics.com/Home.htm Bundled with the Bioptonic 3001 scanner: Bioptonics Viewer] | [http://www.dailymail.co.uk/sciencetech/article-2215052/The-complexity-intricacy-Mother-Nature-revealed-incredible-pictures-plants--seen-inside.html The Daily Mail] | [http://www.guardian.co.uk/science/gallery/2007/sep/04/fruitflybrain#/?picture=330675671&index=1 The Guardian newspaper: 3D Fruit fly] | [http://qt.nokia.com/qt-in-use/ambassadors/project?id=a0F20000006LZ2pEAG Qt Ambassador program] | [http://www.triffidnurseries.co.uk/special3.php Triffid Nurseries website] | |||
<br><br> | |||
|width="50%"| VolViewer is used as a '''stand-alone''' app. or as a '''viewport for other systems''', e.g. Matlab programs. VolViewer uses [http://www.opengl.org/ OpenGL] and [http://qt.nokia.com/products/ Qt] to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. Written by Jerome Avondo it is open-source and is also compatible with the Open Microscopy Environment ([http://openmicroscopy.org/site OME]) (Chris Allen and Avondo, et. al. ''OMERO: flexible, model-driven data management for experimental biology'' Nature Methods 9, 245–253 (2012))<br> [[image:Silique.PNG|360px]]). | |||
|} | |||
==<span style="color:DarkGreen;">Analysing shapes in 2D and 3D: '''AAMToolbox'''== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="10%"| <imgicon>AAMToolbox_logo.jpg|120px|AAMToolbox</imgicon> | |||
|width="40%"|For analysing populations of shapes and colours within the shapes using principal component analysis. <br><br> | |||
[[AAMToolbox Details|'''What? How? Where?''']]<br><br> | |||
[[Tutorials on the Shape modelling toolbox|'''''Tutorials''''': from the beginning]]<br><br> | |||
[[AAMToolbox Download|<span style="color: Gray">'''''Download revised Nov2012''''' </span>]]<br><br> | |||
(PC, Mac, Linux, uses Matlab<br>no Mathworks toolboxes needed<br>[http://www.mathworks.com/products/matlab/tryit.html Matlab 30 day free trial] and <br>[http://www.mathworks.com/academia/student_version/?s_cid=global_nav student edition])<br><br> | |||
|width="50%"| The AAMToolbox enables the user analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading ([http://ieeexplore.ieee.org/xpl/freeabs_all.jsp?arnumber=982900 Matthews ''et al''. 2002][http://www2.cmp.uea.ac.uk/~sjc/matthews-pami-01.pdf version of pdf]), we have used it extensively for analysing the shapes of leaves ([http://www.pnas.org/content/102/29/10221.short Langlade ''et al'' 2005.],[http://www.tandfonline.com/doi/abs/10.2976/1.2836738 Bensmihen ''et al.'' 2010]) and petals ([http://www.sciencemag.org/content/313/5789/963.short Whibley ''et al'' 2006],[http://www.mssaleshops.info/content/21/10/2999.short Feng ''et al''. 2010]). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani. | |||
|} | |||
==<span style="color:DarkGreen;">Analysing the shapes of clones: '''SectorAnalysisToolbox'''== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="10%"| <imgicon>Sector analysis icon.jpg|120px|SectorAnalysisToolbox</imgicon> | |||
|width="40%"|For analysing the shapes of marked cell clones. <br><br> | |||
[[SectorAnalysisToolbox Details|'''What? How? Where?''']]<br><br> | |||
[[SectorAnalysisToolbox Documentation|'''''Tutorials''''': from the beginning]]<br><br> | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSourceDownload_Science_Paper_2012/SectorAnalysisToolbox.zip <span style="color: Gray">'''''Download''''' </span>]<br><br> | |||
(PC, Mac, Linux, uses Matlab<br>no Mathworks toolboxes needed<br>[http://www.mathworks.com/products/matlab/tryit.html Matlab 30 day free trial] and <br>[http://www.mathworks.com/academia/student_version/?s_cid=global_nav student edition])<br><br> | |||
|width="50%"| The SectorAnalysisToolbox enables the user analyse the shapes of marked clones in a sheet of tissue. | |||
|} | |||
=<span style="color:Navy;">'''Algorithms'''= | |||
==<span style="color:Chocolate;">MSERs, extrema, connected-set filters and sieves== | |||
====<span style="color:Chocolate;">The algorithm finding MSER's starts with a connected-set opening or 'o' sieve</span>==== | |||
[[Comparison of Matlab MSER's and 'o' sieve|Comparison of Matlab MSER's and 'o' sieve]] Essentially, no difference. | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
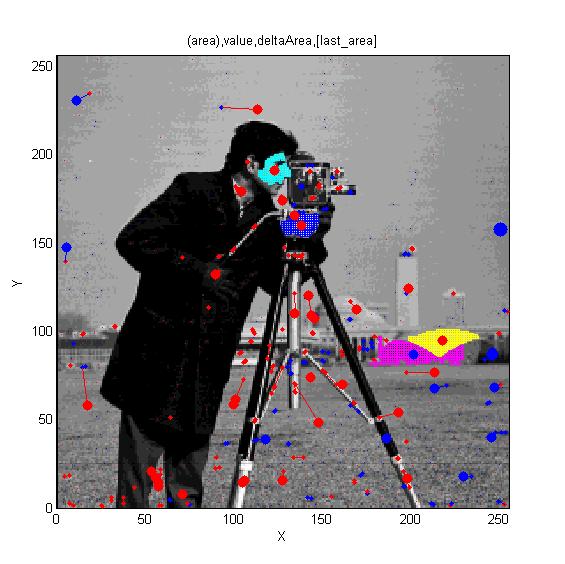
|width="50%"| [[Image:Cameraman_iso_topview.jpg|300px|link=AAMToolbox Details|MSERs]] Cameraman image. Superimposed red spots are maximal extrema and blue spots are minima. Irregular cyan, blue and yellow regions illustrate regions associated with maxima and the magenta region is a minimum. | |||
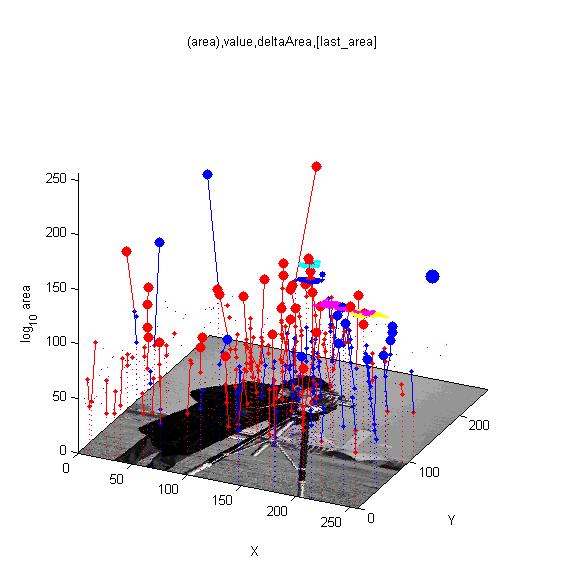
|[[Image:cameraman_iso_tree.jpg|300px|link=AAMToolbox Details|MSERs over scale-space]]<br>Isometric view of the cameraman image with superimposed maxima (red) and minima (blue). The trees trace the maxima through increasing scale-space. Large spots have been identified as stable extrema. | |||
|} | |||
===<span style="color: #C364C5;">Finding interest points, features and segmenting images. </span>=== | |||
#[[MSER and Sieve Details|<span style="color:Chocolate;">'''Technical briefing'''</span>]] <span style="color: #C364C5;">MSER's incorporate 'o' sieves. | |||
#[[MSER's and Connected sets|<span style="color:Chocolate;">'''The twist''': from restricted median filters to sieves and MSER's</span>]] | |||
#One dimensional sieves (measure length) | |||
##[[Types of 1D sieve|Types of 1D sieve]] | |||
##[[First applied to hydrophobicity plots|First applied to hydrophobicity plots]] but lets exploit their idempotency. | |||
#Two dimensional sieves (measure areas) | |||
##Properties | |||
##Relation to MSER's | |||
#Three dimensional sieves (measure volumes) | |||
##[[Segment by volume|Segment by volume]] instant gratification. | |||
<!--[[siv Download|<span style="color: Gray">'''''Download''''' </span>]]<br><br>--> | |||
<!-- [[Software#MSERs extrema connected-set filters and sieves|<span style="color:Green;">'''MORE'''</span>]] --> | |||
===<span style="color:Chocolate;">Art, extrema of light and shade: '''''PhotoArtMaster'''''=== | |||
Art created using ArtMaster, and ArtMaster itself was featured in an exhibit at the London Victoria and Albert (V&A) Museum exhibition <span style="color:Chocolate;">'Cheating? How to make Perfect Work of Art' (2003).</span> The exhibition centered on the idea of [http://en.wikipedia.org/wiki/Hockney%E2%80%93Falco_thesis Hockney's] that advances in realism and accuracy in the history of Western art since the Renaissance were primarily the result of optical aids such as the camera obscura, camera lucida, and curved mirrors. My exhibit used a touch screen (rare in those days) and ArtMaster to help visitors create 'paintings' from photographs. [http://www.sciencemuseum.org.uk/visitmuseum/galleries/turing.aspx finding its name]. (It is entirely different in principle from the software more recently used by Hockney to paint with an iPad.)<br> | |||
{| border="0" width=100% style="background-color:#ffffff;" | |||
|- | |||
|align="center"|[[Image:DegasLightAndShade.jpg|400px]][[Image:Emma_face_Art_C.jpg|300px]] | |||
<br><br> | |||
Illustration of PhotoArtMaster used to find and 'paint' with regions of light and shade crisply segmented from a photograph. Likewise, on the right, edges. | |||
|} | |||
=====PhotoArtMaster===== | |||
Saturday 07/06/2014: Inspired Photographer of the Year 2013 Tony Bennett when asked whether his photograph [http://www.bbc.co.uk/programmes/galleries/p020hd8s Mists and Reflections] had been Photoshopped replied something like <br> | |||
"A digital camera delivers an unemotional ''raw'' image of pixels that you have to manipulate to ''create your photograph''" Photographers manipulate as little as possible. <br> | |||
However there is '''another path one that creates pictures'''. For this you need another piece of software: PhotoArtMaster (ArtMaster). Professional Photographer said ''"'''Forget any comparison whatsoever with the art filters in Photoshop - this software reaches out and enters different stratospheres'''"'' [[Professional Photographer]].<br><br> | |||
Early versions of PhotoArtMaster are still '''available from Amazon''' at low prices (I'm not sure where they come from.) | |||
[http://www.amazon.co.uk/s/ref=nb_sb_noss?url=search-alias%3Daps&field-keywords=photoartmaster] . Some help for both the early versions and the latest version can be found in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this document''''' </span>]). | |||
=====Links to third party PhotoArtMastered pictures===== | |||
*[https://picasaweb.google.com/113257474829608374943/InTheStyleOf Oliver Bangham] Colouful rounded shapes from, yes, my brother. | |||
*[http://en.wikipedia.org/wiki/Wimbledon_%28film%29 The entry sequence of the comedy film 'Wimbledon'] | |||
====The final version of the Windows ArtMaster2.0 [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/OpenSource_ArtMaster/ArtMaster2.0Release.zip <span style="color: #B31B1B">'''''is downloadable here'''''</span>] with no support.==== | |||
Unzip into (for example) the "Program Files" directory then set your system environment to include: ''C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin;'' (You may need help for this. I right clicked 'computer' from the 'Start' menu, then selected 'Advanced system settings', then 'Environment Variables' and finally slid through the System variables until I found and selected 'Path'. This allowed me to edit the path by adding ';C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin' to the end). Rather detailed help using the software is available in [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''this essay''''' </span>].<br><br> | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/Art_For_All_a4a_3_web2.pdf <span style="color: Chocolate">'''''The sieve algorithms underpinning PhotoArtMaster software''''' </span>] are described in an extract of the | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/SieveWebPages/a4a_2_screensize.pdf <span style="color: Chocolate">''''essay''''' </span>]. These documents were written to support our Fo2Pix company. PhotoArtMaster originally sold >65,000 licences but ill health forced the closure of Fo2Pix. | |||
{| border="0" width=100% style="background-color:#ffffff;" | |||
|- | |||
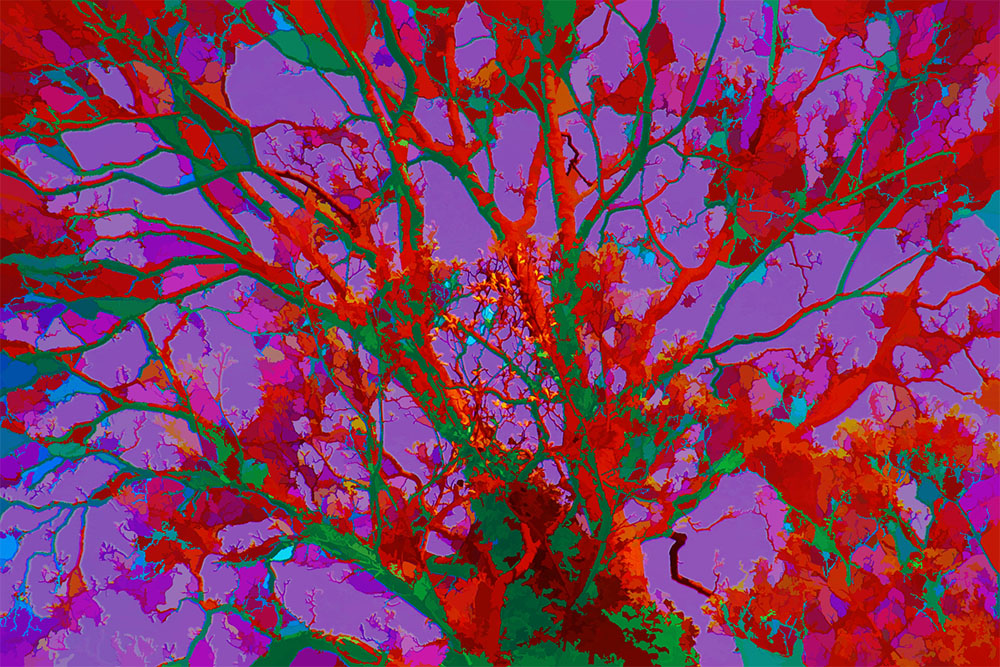
|align="center"|[[Image:Trees_1.jpg|600px]] | |||
<br><br> | |||
Illustration of PhotoArtMaster used to find and 'paint' with regions of colour crisply segmented from a photograph. | |||
|} | |||
<!-- [[Documentation_of_Connected_Set_Filters_or_Sieves|Art test page]]<br><br> | |||
[[Documentation_of_Connected_Set_Filters_or_Sieves]] --> | |||
==<span style="color:Navy;">Reaction-diffusion and morphogenesis== | |||
{| border="0" width=100% style="background-color:#000000;" | |||
|- | |||
|align="center"| | |||
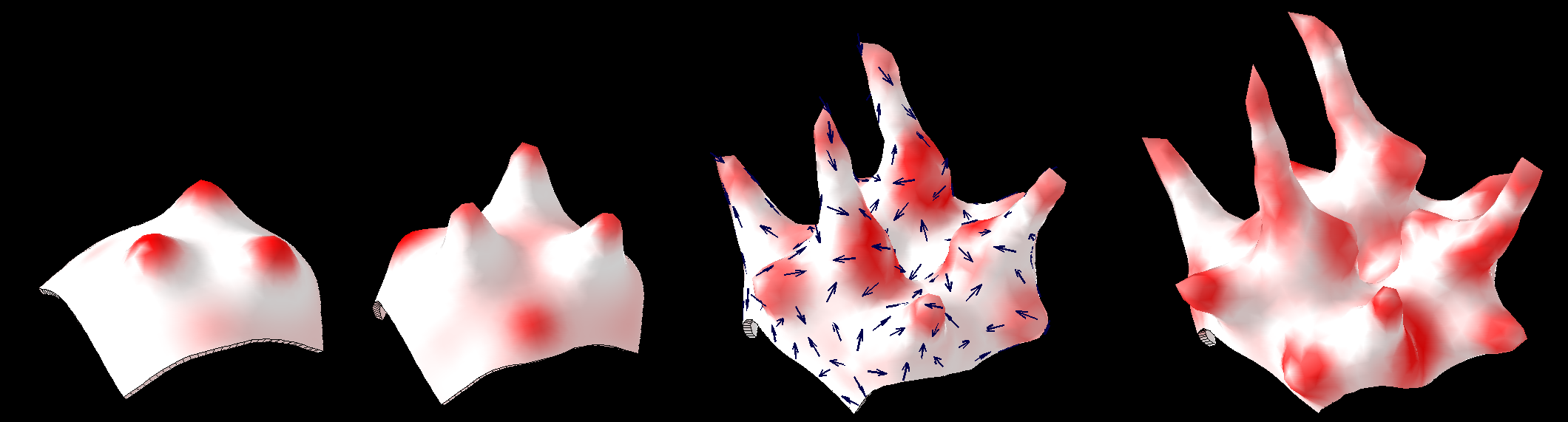
[[Image:tentacles_morphogenesis.png|600px]] | |||
|} | |} | ||
Illustration of morphogenesis inspired by Turing's paper. <br><br> | |||
[http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/GPT_ReactionDiffusionTentacles_20121211.zip <span style="color: Navy">Example using growth toolbox GPT_ReactionDiffusionTentacles_20121211.zip</span>] | |||
<br><br> | |||
===1 A=== | |||
=<span style="color:DarkGreen;">Open source systems to which we | =<span style="color:DarkGreen;">Open source systems to which we have contributed= | ||
==<span style="color:DarkGreen;">OMERO== | ==<span style="color:DarkGreen;">OMERO== | ||
{| border="0" cellpadding="5" cellspacing="5" | {| border="0" cellpadding="5" cellspacing="5" | ||
|- valign="top" | |- valign="top" | ||
|width="10%"| <imgicon>OMERO_DIAGRAM.jpg|100px|OMERO</imgicon> | |width="10%"| <imgicon>OMERO_DIAGRAM.jpg|100px|OMERO</imgicon> | ||
|width="40%"|For working with the OME image database. <br><br>See [http://www.openmicroscopy.org/site/products/omero ''Details''], [http://www.openmicroscopy.org/site/support/omero4/downloads ''Download'']<br><br> | |width="40%"|For working with the OME image database. <br><br>See [http://www.openmicroscopy.org/site/products/omero ''Details''], [http://www.openmicroscopy.org/site/support/omero4/downloads ''Download'']<br> [http://dmbi.nbi.bbsrc.ac.uk/index.php/OMEROWorkshop OMERO Workshop] <br><br> | ||
(Windows, Mac, Linux) | (Windows, Mac, Linux) | ||
|width="50%"| [http://openmicroscopy.org/site/support/omero4 Open Microscopy Environment Remote Objects (OMERO).] for visualising, managing, and annotating scientific image data. See also our [http://dmbi.nbi.bbsrc.ac.uk/index.php/OMEROWorkshop OMERO Workshop] training course we ran in April 2011. | |width="50%"| [http://openmicroscopy.org/site/support/omero4 Open Microscopy Environment Remote Objects (OMERO).] for visualising, managing, and annotating scientific image data. See also our [http://dmbi.nbi.bbsrc.ac.uk/index.php/OMEROWorkshop OMERO Workshop] training course we ran in April 2011. | ||
|} | |} | ||
=<span style="color:DarkGreen;">Tools and Utilities= | =<span style="color:DarkGreen;">Tools and Utilities= | ||
| Line 42: | Line 171: | ||
|- valign="top" | |- valign="top" | ||
|width="10%"| <imgicon>BioformatsConverterZip.png|100px|BioformatsConverter</imgicon> | |width="10%"| <imgicon>BioformatsConverterZip.png|100px|BioformatsConverter</imgicon> | ||
|width="40%"|For converting microscope manufacturer proprietary file formats. <br><br>See [[BioformatsConverter|''Details | |width="40%"|For converting microscope manufacturer proprietary file formats. <br><br>See [[BioformatsConverter|''Details'']]<br><br> | ||
(Windows, Mac, Linux) | (Windows, Mac, Linux) | ||
|width="50%"| This tool allows for the batch conversion of microscope manufacturer proprietary file formats, to the open source OME-TIFF standard. Uses the [http://www.loci.wisc.edu/software/bio-formats Bioformats] library. | |width="50%"| This tool allows for the batch conversion of microscope manufacturer proprietary file formats, to the open source OME-TIFF standard. Uses the [http://www.loci.wisc.edu/software/bio-formats Bioformats] library. | ||
|} | |} | ||
==<span style="color:DarkGreen;">Dependency Checking Tool== | |||
Tool for recursively finding what further functions a function depends on. See [[myDepFun|''Details'']] | |||
==<span style="color:DarkGreen;">PIN Point== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="10%"|[[File:PinPoint.jpg]] | |||
|width="40%"|Tool for analysing PIN Signal in cells. | |||
|width="50%"| [http://cmpdartsvr1.cmp.uea.ac.uk/downloads/software/PinPoint.zip <span style="color: Gray">'''''Download''''' </span>]<br><br> | |||
Sample Project | |||
|} | |||
=<span style="color:DarkGreen;">In development</span>= | |||
==<span style="color:DarkGreen;">MTtbox</span>== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="10%"| <imgicon>MTtboxA.jpg|100px|BioformatsConverter</imgicon> | |||
|width="40%"|For modelling the behaviour of microtubules within a cell. <br><br> | |||
See [[MTtbox documentation|''Details'']]<br><br> | |||
(Windows, Mac, Linux) | |||
|width="50%"| In development. The idea is to be able to model the behaviour of growing microtubules and factors as they react chemically and diffuse within the different cell compartments.<br><br> | |||
The icon shows a spherical cell sliced open to show concentric components: cell wall (magenta), plasma-membrane (yellow), cytoplasm (green) and vacuole (yellow). Microtubules (blue) grow in 3D within the cytoplasm. | |||
|} | |||
=<span style="color:Navy;">Historical</span>= | |||
==<span style="color:Navy;">Robot arm: still in production after 30 years (serving local industry)</span>== | |||
{| border="0" cellpadding="5" cellspacing="5" | |||
|- valign="top" | |||
|width="50%"|[[Image:2014-06-10 14.22.20 small robot.jpg|400px|]] | |||
|width="40%"|For teaching '''production control''' and '''interrupt programming'''. <br><br> | |||
1983 and we are in a world of '''Apple II and BBC B computers''' - the ''6502'' processor reigns. Particularly good for real-time control it responded very fast to hardware interrupts from, for example, the timer. To illustrate timer interrupts what better than digital servo-motors? Set up the on-board timers to produce a stream of 'heartbeat' of pulses, one every 20 ms out of the parallel port and control up to eight motors. Pulse widths, from 1 to 2 ms, control the position of each motor arm. Derek Fulton and I made some loose lab. money by writing a series of articles showing exactly how to build and, in particular, control this robot arm.[[Publications#.28G.29_Computer_control.2C_measurement_and_commercial_software |Bangham et al.]]. | |||
Our copyright, I took it to a local company LJ-Electronics ([http://www.ljcreate.com/products/product.asp?id=314&program=195&curr=2 now LJ-Creative]) who incorporated it detail for detail into their product line. Originally, they called it the Emu. Still in production: '''Lovely outcome.''' | |||
|} | |||
=Historical= | |||
Latest revision as of 10:49, 10 December 2021
Current activity: a collaboration with the CoenLab with the aim of understanding how patterns of gene activity in biological organs influence the developing shape. The BanghamLab is focussed on the conceptual underpinning: concepts captured in computational growth models, experimental data visualisation and analysis.
Notes on documenting our software
Notes for Lab members on how to contribute to this Wiki and where to put downloads.
Matlab tip: searching a large data structure for a particular field. Clear the command window. Evaluate the structure to list all the fields, then use the usual control-f search tool on the command window.
Computational biology
Quantitative understanding of growing shapes: GFtbox
We developed GFtbox to allow us to model the growth of complex shapes with the ultimate goal: to understand the relationships between genes, growth and form.
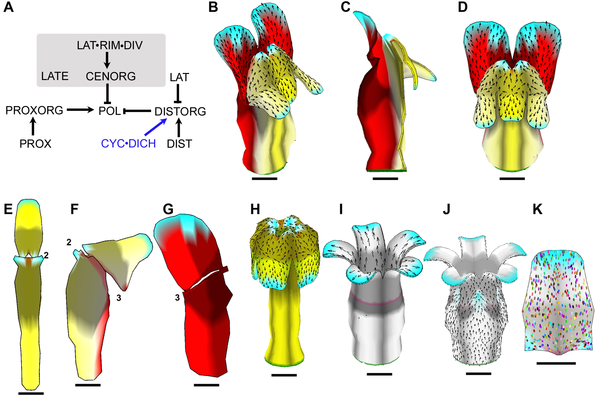 Example of a growing snapdragon flower and some mutants ( Green et al 2011). Growth is specified by factors (genes) according to the Growing Polarised Tissue Framework. Colours represent putative gene activity, arrows the polariser gradient and spots clones. Example of a growing snapdragon flower and some mutants ( Green et al 2011). Growth is specified by factors (genes) according to the Growing Polarised Tissue Framework. Colours represent putative gene activity, arrows the polariser gradient and spots clones.
|
GPT_Snapdragon_2010_Green_et_al-0002.png</wikiflv> |
| <imgicon>GPT_thumbnail2.png|120px|GFtbox</imgicon> |
For modelling the growth of shapes. Ready Reference Manual
What? How? Where? Background |
Viewing and measuring volume images: VolViewer
| <imgicon>VolViewer-logo.png|120px|VolViewer</imgicon> | For viewing and measuring volume images on both normal and stereo screens. Typical images from: confocal microscope and Optical Projection Tomography (OPT) images What? How? Where? Cell: Online Gallery | Front cover: Handbook of Plant Science | Front cover: The Plant Cell | American Scientist | Royal Microscopical Society: Infocus Magazine | Bundled with the Bioptonic 3001 scanner: Bioptonics Viewer | The Daily Mail | The Guardian newspaper: 3D Fruit fly | Qt Ambassador program | Triffid Nurseries website
|
VolViewer is used as a stand-alone app. or as a viewport for other systems, e.g. Matlab programs. VolViewer uses OpenGL and Qt to provide a user friendly application to interactively explore and quantify multi-dimensional biological images. It has been successfully used in our lab to explore and quantify confocal microscopy and optical projection tomography images. Written by Jerome Avondo it is open-source and is also compatible with the Open Microscopy Environment (OME) (Chris Allen and Avondo, et. al. OMERO: flexible, model-driven data management for experimental biology Nature Methods 9, 245–253 (2012)) ). ).
|
Analysing shapes in 2D and 3D: AAMToolbox
| <imgicon>AAMToolbox_logo.jpg|120px|AAMToolbox</imgicon> | For analysing populations of shapes and colours within the shapes using principal component analysis. What? How? Where?
|
The AAMToolbox enables the user analyse the shape and colour of collections of similar objects. Originally developed to analyse face shapes for lipreading (Matthews et al. 2002version of pdf), we have used it extensively for analysing the shapes of leaves (Langlade et al 2005.,Bensmihen et al. 2010) and petals (Whibley et al 2006,Feng et al. 2010). The analysis can be applied to art, for example, finding systematic differences between portraits by Rembrandt and Modigliani. |
Analysing the shapes of clones: SectorAnalysisToolbox
| <imgicon>Sector analysis icon.jpg|120px|SectorAnalysisToolbox</imgicon> | For analysing the shapes of marked cell clones. What? How? Where? |
The SectorAnalysisToolbox enables the user analyse the shapes of marked clones in a sheet of tissue. |
Algorithms
MSERs, extrema, connected-set filters and sieves
The algorithm finding MSER's starts with a connected-set opening or 'o' sieve
Comparison of Matlab MSER's and 'o' sieve Essentially, no difference.
Finding interest points, features and segmenting images.
- Technical briefing MSER's incorporate 'o' sieves.
- The twist: from restricted median filters to sieves and MSER's
- One dimensional sieves (measure length)
- Types of 1D sieve
- First applied to hydrophobicity plots but lets exploit their idempotency.
- Two dimensional sieves (measure areas)
- Properties
- Relation to MSER's
- Three dimensional sieves (measure volumes)
- Segment by volume instant gratification.
Art, extrema of light and shade: PhotoArtMaster
Art created using ArtMaster, and ArtMaster itself was featured in an exhibit at the London Victoria and Albert (V&A) Museum exhibition 'Cheating? How to make Perfect Work of Art' (2003). The exhibition centered on the idea of Hockney's that advances in realism and accuracy in the history of Western art since the Renaissance were primarily the result of optical aids such as the camera obscura, camera lucida, and curved mirrors. My exhibit used a touch screen (rare in those days) and ArtMaster to help visitors create 'paintings' from photographs. finding its name. (It is entirely different in principle from the software more recently used by Hockney to paint with an iPad.)
 
|
PhotoArtMaster
Saturday 07/06/2014: Inspired Photographer of the Year 2013 Tony Bennett when asked whether his photograph Mists and Reflections had been Photoshopped replied something like
"A digital camera delivers an unemotional raw image of pixels that you have to manipulate to create your photograph" Photographers manipulate as little as possible.
However there is another path one that creates pictures. For this you need another piece of software: PhotoArtMaster (ArtMaster). Professional Photographer said "Forget any comparison whatsoever with the art filters in Photoshop - this software reaches out and enters different stratospheres" Professional Photographer.
Early versions of PhotoArtMaster are still available from Amazon at low prices (I'm not sure where they come from.)
[1] . Some help for both the early versions and the latest version can be found in 'this document ).
Links to third party PhotoArtMastered pictures
- Oliver Bangham Colouful rounded shapes from, yes, my brother.
- The entry sequence of the comedy film 'Wimbledon'
The final version of the Windows ArtMaster2.0 is downloadable here with no support.
Unzip into (for example) the "Program Files" directory then set your system environment to include: C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin; (You may need help for this. I right clicked 'computer' from the 'Start' menu, then selected 'Advanced system settings', then 'Environment Variables' and finally slid through the System variables until I found and selected 'Path'. This allowed me to edit the path by adding ';C:\Program Files\Pam2.0 Release\jre\bin;C:\Program Files\Pam2.0 Release\bin' to the end). Rather detailed help using the software is available in 'this essay .
The sieve algorithms underpinning PhotoArtMaster software are described in an extract of the
'essay . These documents were written to support our Fo2Pix company. PhotoArtMaster originally sold >65,000 licences but ill health forced the closure of Fo2Pix.
|
Reaction-diffusion and morphogenesis
Illustration of morphogenesis inspired by Turing's paper.
Example using growth toolbox GPT_ReactionDiffusionTentacles_20121211.zip
1 A
Open source systems to which we have contributed
OMERO
| <imgicon>OMERO_DIAGRAM.jpg|100px|OMERO</imgicon> | For working with the OME image database. See Details, Download OMERO Workshop (Windows, Mac, Linux) |
Open Microscopy Environment Remote Objects (OMERO). for visualising, managing, and annotating scientific image data. See also our OMERO Workshop training course we ran in April 2011. |
Tools and Utilities
BioformatsConverter
| <imgicon>BioformatsConverterZip.png|100px|BioformatsConverter</imgicon> | For converting microscope manufacturer proprietary file formats. See Details (Windows, Mac, Linux) |
This tool allows for the batch conversion of microscope manufacturer proprietary file formats, to the open source OME-TIFF standard. Uses the Bioformats library. |
Dependency Checking Tool
Tool for recursively finding what further functions a function depends on. See Details
PIN Point
|
Tool for analysing PIN Signal in cells. | Download Sample Project |
In development
MTtbox
| <imgicon>MTtboxA.jpg|100px|BioformatsConverter</imgicon> | For modelling the behaviour of microtubules within a cell. See Details |
In development. The idea is to be able to model the behaviour of growing microtubules and factors as they react chemically and diffuse within the different cell compartments. The icon shows a spherical cell sliced open to show concentric components: cell wall (magenta), plasma-membrane (yellow), cytoplasm (green) and vacuole (yellow). Microtubules (blue) grow in 3D within the cytoplasm. |
Historical
Robot arm: still in production after 30 years (serving local industry)

|
For teaching production control and interrupt programming. 1983 and we are in a world of Apple II and BBC B computers - the 6502 processor reigns. Particularly good for real-time control it responded very fast to hardware interrupts from, for example, the timer. To illustrate timer interrupts what better than digital servo-motors? Set up the on-board timers to produce a stream of 'heartbeat' of pulses, one every 20 ms out of the parallel port and control up to eight motors. Pulse widths, from 1 to 2 ms, control the position of each motor arm. Derek Fulton and I made some loose lab. money by writing a series of articles showing exactly how to build and, in particular, control this robot arm.Bangham et al.. Our copyright, I took it to a local company LJ-Electronics (now LJ-Creative) who incorporated it detail for detail into their product line. Originally, they called it the Emu. Still in production: Lovely outcome. |